7VZ2
 
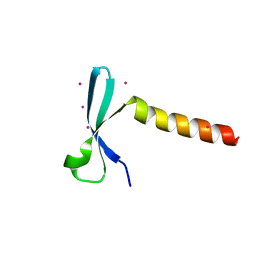 | | Crystal structure of chromodomain of Arabidopsis LHP1 | | Descriptor: | Chromo domain-containing protein LHP1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Min, J. | | Deposit date: | 2021-11-15 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the recognition of methylated histone H3 by the Arabidopsis LHP1 chromodomain.
J.Biol.Chem., 298, 2022
|
|
3QO2
 
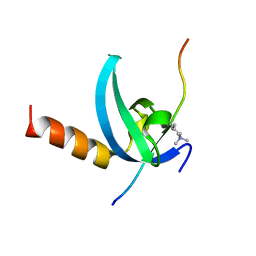 | | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9 | | Descriptor: | 1,2-ETHANEDIOL, Histone H3 peptide, M-phase phosphoprotein 8 | | Authors: | Chang, Y, Horton, J.R, Bedford, M.T, Zhang, X, Cheng, X. | | Deposit date: | 2011-02-09 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding.
J.Mol.Biol., 408, 2011
|
|
5YY9
 
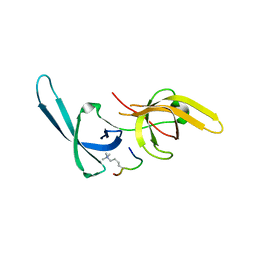 | | Crystal structure of Tandem Tudor Domain of human UHRF1 in complex with LIG1-K126me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Ligase 1 | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
5YYA
 
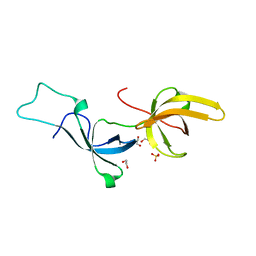 | | Crystal structure of Tandem Tudor Domain of human UHRF1 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
6A5M
 
 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 2 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5N
 
 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5K
 
 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 1 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ASZ
 
 | |
6AT0
 
 | |
6C23
 
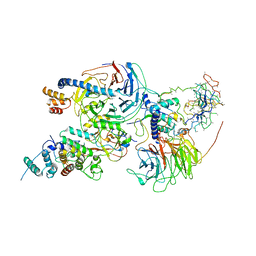 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-05 | | Release date: | 2018-01-24 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
6C24
 
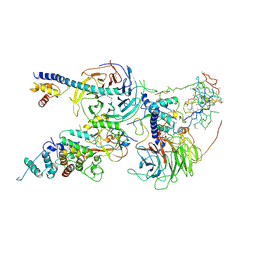 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-06 | | Release date: | 2018-01-24 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
