9CDE
 
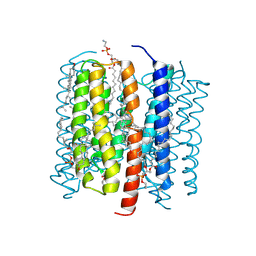 | | Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Continuous Illumination State | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Kalium Channelrhodopsin 1 C110A - Continuous Illumination State, ... | | Authors: | Morizumi, T, Kim, K, Ernst, O.P. | | Deposit date: | 2024-06-24 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural insights into light-gating of potassium-selective channelrhodopsin.
Nat Commun, 16, 2025
|
|
9CDD
 
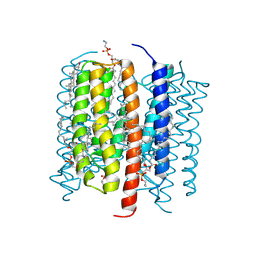 | | Kalium channelrhodopsin 1 C110A mutant from Hyphochytrium catenoides, Laser-Flash-Illuminated | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Kalium Channelrhodopsin 1, ... | | Authors: | Morizumi, T, Kim, K, Ernst, O.P. | | Deposit date: | 2024-06-24 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into light-gating of potassium-selective channelrhodopsin.
Nat Commun, 16, 2025
|
|
4XF5
 
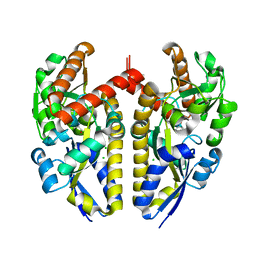 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound (S)-(+)-2-Amino-1-propanol. | | Descriptor: | (2S)-2-aminopropan-1-ol, CHLORIDE ION, Twin-arginine translocation pathway signal | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-26 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound (S)-(+)-2-Amino-1-propanol.
To be published
|
|
4XAL
 
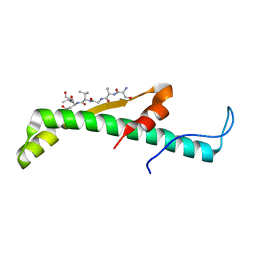 | |
6QXD
 
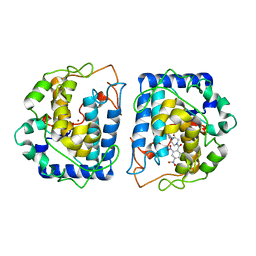 | | Crystal Structure of tyrosinase from Bacillus megaterium with JKB inhibitor in the active site. | | Descriptor: | (2,4-dinitrophenyl)-[4-[(4-fluorophenyl)methyl]piperazin-1-yl]methanone, COPPER (II) ION, Tyrosinase | | Authors: | Deri Zenaty, B, Gitto, R, Pazy, Y, Fishman, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.317 Å) | | Cite: | Exploiting the 1-(4-fluorobenzyl)piperazine fragment for the development of novel tyrosinase inhibitors as anti-melanogenic agents: Design, synthesis, structural insights and biological profile.
Eur.J.Med.Chem., 178, 2019
|
|
6WF2
 
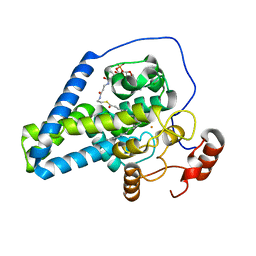 | | Crystal structure of mouse SCD1 with a diiron center | | Descriptor: | Acyl-CoA desaturase 1, FE (III) ION, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Shen, J, Zhou, M. | | Deposit date: | 2020-04-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure and Mechanism of a Unique Diiron Center in Mammalian Stearoyl-CoA Desaturase.
J.Mol.Biol., 432, 2020
|
|
4XAR
 
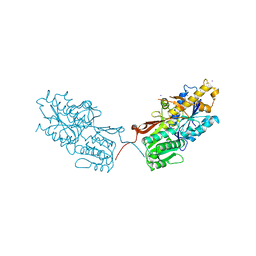 | | mGluR2 ECD and mGluR3 ECD complex with ligands | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, IODIDE ION, Metabotropic glutamate receptor 3 | | Authors: | Clawson, D.K. | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-Disubstituted Analogs of 1S,2S,5R,6S-2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylate: Identification of a Potent, Selective Metabotropic Glutamate Receptor Agonist and Determination of Agonist-Bound Human mGlu2 and mGlu3 Amino Terminal Domain Structures.
J.Med.Chem., 58, 2015
|
|
4XMP
 
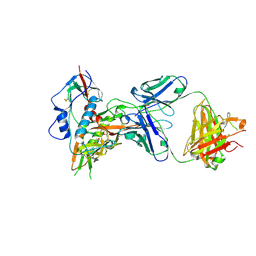 | | Crystal structure of broadly and potently neutralizing antibody VRC08 in complex with HIV-1 clade A strain Q842.d12 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160,Envelope glycoprotein gp160,Envelope glycoprotein gp160, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-01-15 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7831 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell, 161, 2015
|
|
6WI7
 
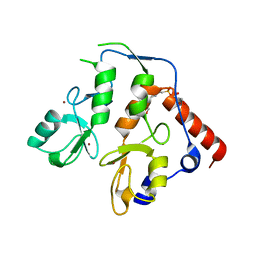 | | RING1B-BMI1 fusion in closed conformation | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase RING2, Polycomb complex protein BMI-1 chimera, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Small-molecule inhibitors targeting Polycomb repressive complex 1 RING domain.
Nat.Chem.Biol., 17, 2021
|
|
8G6K
 
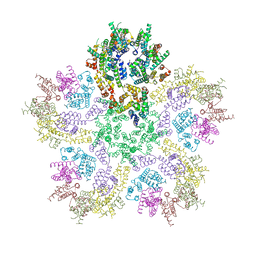 | |
8G6O
 
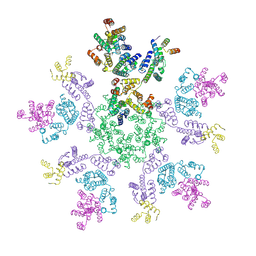 | | HIV-1 capsid lattice bound to IP6 and Lenacapavir | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ZEN
 
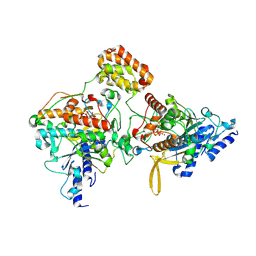 | | Cryo-EM structure of Adr-2-Adbp-1-dsRBD0 complex | | Descriptor: | Adr-2-binding protein 1, Double-stranded RNA-specific adenosine deaminase adr-2, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Liu, Z.M, Mu, J.Q, Wu, C. | | Deposit date: | 2024-05-06 | | Release date: | 2025-05-14 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of Adr-2-Adbp-1-dsRBD0 complex
To Be Published
|
|
8G6L
 
 | | HIV-1 capsid lattice bound to IP6, pH 6.2 | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G6M
 
 | | HIV-1 CA lattice bound to IP6, pH 7.4 | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G6N
 
 | | HIV-1 capsid lattice bound to dNTPs | | Descriptor: | Capsid protein | | Authors: | Highland, C.M, Dick, R.A. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4XJP
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with an inhibitor optimized from HTS lead | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[4-(1,3-benzodioxol-5-ylcarbonyl)piperazin-1-yl]phenyl}ethanone, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, ... | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2015-01-08 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Pyridoxal 5'-Phosphate-Dependent Transaminase Enzyme (BioA) Inhibitors that Target Biotin Biosynthesis in Mycobacterium tuberculosis.
J. Med. Chem., 60, 2017
|
|
8DEI
 
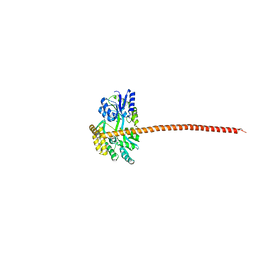 | | Structure of the Cac1 KER domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Maltodextrin-binding protein,Chromatin assembly factor 1 subunit p90 fusion, ... | | Authors: | Rosas, R, Churchill, M.E.A. | | Deposit date: | 2022-06-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A novel single alpha-helix DNA-binding domain in CAF-1 promotes gene silencing and DNA damage survival through tetrasome-length DNA selectivity and spacer function.
Elife, 12, 2023
|
|
4XWP
 
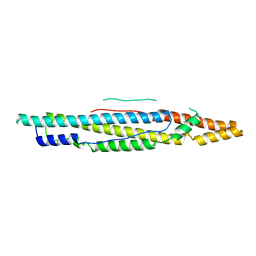 | |
7NWN
 
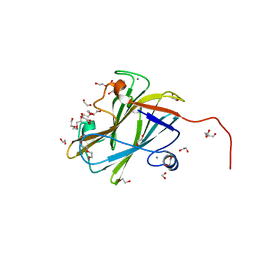 | | A carbohydrate binding module family 9 (CBM9) from Caldicellulsiruptor kristjanssonii | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylanase, CALCIUM ION, ... | | Authors: | Krska, D, Mazurkewich, S, Navarro Poulsen, J, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Functional Analysis of a Multimodular Hyperthermostable Xylanase-Glucuronoyl Esterase from Caldicellulosiruptor kristjansonii .
Biochemistry, 60, 2021
|
|
4XY3
 
 | | Structure of ESX-1 secreted protein EspB | | Descriptor: | ESX-1 secretion-associated protein EspB | | Authors: | Wagner, J.M, Korotkov, K.V. | | Deposit date: | 2015-02-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of EspB, a secreted substrate of the ESX-1 secretion system of Mycobacterium tuberculosis.
J.Struct.Biol., 191, 2015
|
|
4XPO
 
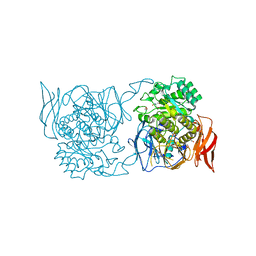 | | Crystal structure of a novel alpha-galactosidase from Pedobacter saltans | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-glucosidase | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
8RP5
 
 | | Alpha-Methylacyl-CoA racemase from Mycobacterium tuberculosis (E241A mutant) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-methylacyl-CoA racemase, TRIETHYLENE GLYCOL | | Authors: | Mojanaga, O.O, Acharya, K.R, Lloyd, M.D. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | alpha-Methylacyl-CoA Racemase from Mycobacterium tuberculosis -Detailed Kinetic and Structural Characterization of the Active Site.
Biomolecules, 14, 2024
|
|
8RP4
 
 | | Alpha-Methylacyl-CoA racemase from Mycobacterium tuberculosis (D156A mutant) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-methylacyl-CoA racemase, DI(HYDROXYETHYL)ETHER | | Authors: | Mojanaga, O.O, Acharya, K.R, Lloyd, M.D. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | alpha-Methylacyl-CoA Racemase from Mycobacterium tuberculosis -Detailed Kinetic and Structural Characterization of the Active Site.
Biomolecules, 14, 2024
|
|
6OYD
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-004 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-05-14 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
7NWO
 
 | | A carbohydrate binding module family 9 (CBM9) from Caldicellulosiruptor kristjanssonii in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylanase, CALCIUM ION, ... | | Authors: | Krska, D, Mazurkewich, S, Navarro Poulsen, J, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of a Multimodular Hyperthermostable Xylanase-Glucuronoyl Esterase from Caldicellulosiruptor kristjansonii .
Biochemistry, 60, 2021
|
|
