6Q8Y
 
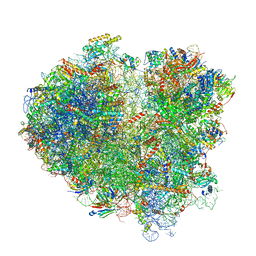 | | Cryo-EM structure of the mRNA translating and degrading yeast 80S ribosome-Xrn1 nuclease complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Heckel, E, Cheng, J, Buschauer, R, Kater, L, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2018-12-16 | | Release date: | 2019-03-13 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the 80S ribosome-Xrn1 nuclease complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2V0W
 
 | |
8FON
 
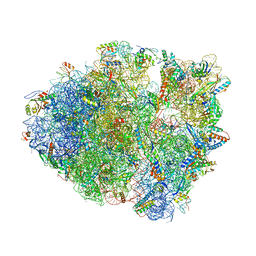 | | Crystal structure of tRNA^Lys(SUU) bound to AUA codon in the ribosomal P site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2023-01-02 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structural basis for reduced ribosomal A-site fidelity in response to P-site codon-anticodon mismatches.
J.Biol.Chem., 299, 2023
|
|
4V1N
 
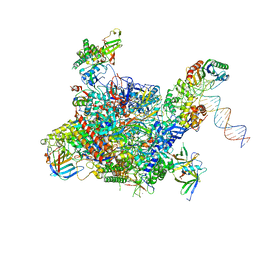 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
4V4J
 
 | | Interactions and Dynamics of the Shine-Dalgarno Helix in the 70S Ribosome. | | Descriptor: | 16S RNA, 23S LARGE SUBUNIT RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Trakhanov, S, Asahara, H, Laurberg, M, Noller, H.F. | | Deposit date: | 2007-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.83 Å) | | Cite: | Interactions and dynamics of the Shine Dalgarno helix in the 70S ribosome.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4V67
 
 | | Crystal structure of a translation termination complex formed with release factor RF2. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Asahara, H, Lancaster, L, Laurberg, M, Hirschi, A, Noller, H.F. | | Deposit date: | 2008-10-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a translation termination complex formed with release factor RF2.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4V1O
 
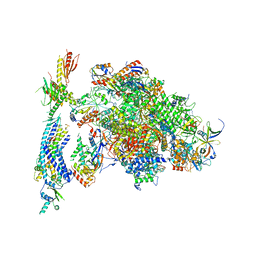 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
1DGO
 
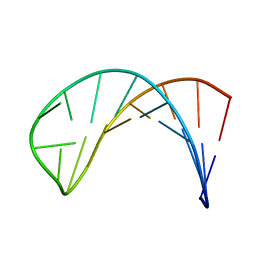 | |
5U5C
 
 | | Coiled Coil Peptide Metal Coordination Framework: Tetramer Fold | | Descriptor: | COPPER (II) ION, Designed tetrameric coiled coil peptide with one terpyridine side chain, HEXANE-1,6-DIOL | | Authors: | Tavenor, N.A, Murnin, M.J, Horne, W.S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Supramolecular Metal-Coordination Polymers, Nets, and Frameworks from Synthetic Coiled-Coil Peptides.
J. Am. Chem. Soc., 139, 2017
|
|
5TLA
 
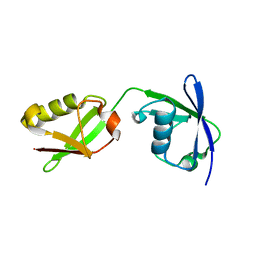 | |
1S1K
 
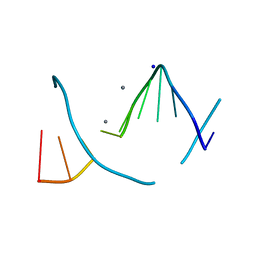 | |
1SGI
 
 | | Crystal structure of the anticoagulant slow form of thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, thrombin | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
7S5S
 
 | | CTX-M-15 WT in complex with BLIP WT | | Descriptor: | Beta-lactamase, Beta-lactamase inhibitory protein | | Authors: | Lu, S, Palzkill, T, Hu, L.Y, Prasad, B.V.V, Sankaran, B. | | Deposit date: | 2021-09-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An active site loop toggles between conformations to control antibiotic hydrolysis and inhibition potency for CTX-M beta-lactamase drug-resistance enzymes.
Nat Commun, 13, 2022
|
|
1SA4
 
 | | human protein farnesyltransferase complexed with FPP and R115777 | | Descriptor: | 6-[(S)-AMINO(4-CHLOROPHENYL)(1-METHYL-1H-IMIDAZOL-5-YL)METHYL]-4-(3-CHLOROPHENYL)-1-METHYLQUINOLIN-2(1H)-ONE, FARNESYL DIPHOSPHATE, Protein farnesyltransferase beta subunit, ... | | Authors: | Reid, T.S, Beese, L.S. | | Deposit date: | 2004-02-06 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Anticancer Clinical Candidates R115777 (Tipifarnib) and BMS-214662 Complexed with Protein Farnesyltransferase Suggest a Mechanism of FTI Selectivity.
Biochemistry, 43, 2004
|
|
1S64
 
 | | Rat protein geranylgeranyltransferase type-I complexed with L-778,123 and a sulfate anion | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[(5-{[4-(3-CHLOROPHENYL)-3-OXOPIPERAZIN-1-YL]METHYL}-1H-IMIDAZOL-1-YL)METHYL]BENZONITRILE, CHLORIDE ION, ... | | Authors: | Reid, T.S, Long, S.B, Beese, L.S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic Analysis Reveals that Anticancer Clinical Candidate L-778,123 Inhibits Protein Farnesyltransferase and Geranylgeranyltransferase-I by Different Binding Modes.
Biochemistry, 43, 2004
|
|
7SWY
 
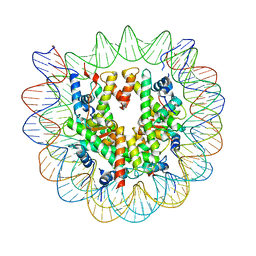 | | 2.6 A structure of a 40-601[TA-rich+1]-40 nucleosome | | Descriptor: | DNA Guide Strand, DNA Tracking Strand, Histone H2A, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Armache, J.-P. | | Deposit date: | 2021-11-21 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5J0N
 
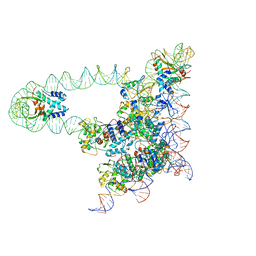 | | Lambda excision HJ intermediate | | Descriptor: | Excisionase, Integrase, Integration host factor subunit alpha, ... | | Authors: | Van Duyne, G, Grigorieff, N, Landy, A. | | Deposit date: | 2016-03-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structure of a Holliday junction complex reveals mechanisms governing a highly regulated DNA transaction.
Elife, 5, 2016
|
|
5J3C
 
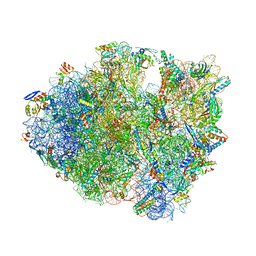 | |
5VBF
 
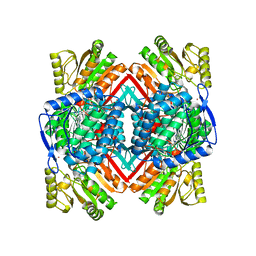 | |
5VYC
 
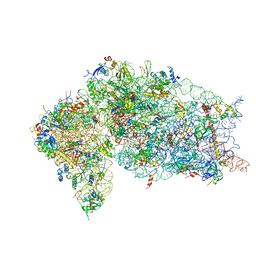 | | Crystal structure of the human 40S ribosomal subunit in complex with DENR-MCT-1. | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Lomakin, I.B, Stolboushkina, E.A, Vaidya, A.T, Garber, M.B, Dmitriev, S.E, Steitz, T.A. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal Structure of the Human Ribosome in Complex with DENR-MCT-1.
Cell Rep, 20, 2017
|
|
4U8U
 
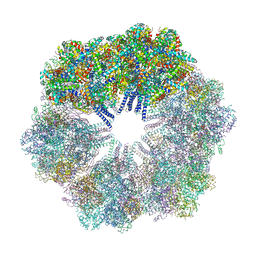 | | The Crystallographic structure of the giant hemoglobin from Glossoscolex paulistus at 3.2 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Bachega, J.F.R, Maluf, F.V, Andi, B, D'Muniz Pereira, H, Carazzollea, M.F, Orville, A, Tabak, M, Garratt, R.C, Horjales, E. | | Deposit date: | 2014-08-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the giant haemoglobin from Glossoscolex paulistus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2C12
 
 | | Crystal Structure of Nitroalkane Oxidase in Complex with Spermine, a Competitive Inhibitor | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Nagpal, A, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2005-09-10 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structures of Nitroalkane Oxidase: Insights Into the Reaction Mechanism from a Covalent Complex of the Flavoenzyme Trapped During Turnover.
Biochemistry, 45, 2006
|
|
6WP0
 
 | |
6L4U
 
 | | Structure of the PSI-FCPI supercomplex from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Miyazaki, N, Akita, F, Shen, J.R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of photosystem I-light-harvesting supercomplex from a red-lineage diatom
Nat Commun, 2020
|
|
6L4T
 
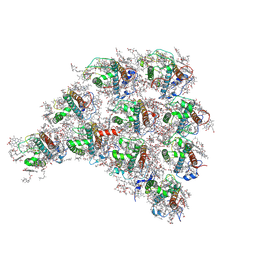 | | Structure of the peripheral FCPI from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Miyazaki, N, Akita, F, Shen, J.R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of photosystem I-light-harvesting supercomplex from a red-lineage diatom
Nat Commun, 2020
|
|
