8DTU
 
 | | The complex of nanobody 5344N74D with BCL11A ZF6. | | Descriptor: | B-cell lymphoma/leukemia 11A, Nanobody 5344N74D, ZINC ION | | Authors: | Yin, M, Tenglin, K, Zhai, L, Dassama, L.M, Orkin, S.H. | | Deposit date: | 2022-07-26 | | Release date: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Evolution of nanobodies specific for BCL11A.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E15
 
 | | A computationally stabilized hMPV F protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F1 protein with Fibritin peptide, F2 protein, ... | | Authors: | Huang, J, Gonzalez, K, Mousa, J, Strauch, E. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A general computational design strategy for stabilizing viral class I fusion proteins.
Nat Commun, 15, 2024
|
|
6ALK
 
 | | NMR solution structure of the major beech pollen allergen Fag s 1 | | Descriptor: | Fag s 1 pollen allergen | | Authors: | Moraes, A.H, Asam, A, Almeida, F.C.L, Wallner, M, Ferreira, F, Valente, A.P. | | Deposit date: | 2017-08-08 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for cross-reactivity and conformation fluctuation of the major beech pollen allergen Fag s 1.
Sci Rep, 8, 2018
|
|
7EA6
 
 | | Crystal structure of TCR-017 ectodomain | | Descriptor: | T cell receptor 017 alpha chain, T cell receptor 017 beta chain | | Authors: | Nagae, M, Yamasaki, S. | | Deposit date: | 2021-03-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18000245 Å) | | Cite: | Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients.
J.Exp.Med., 218, 2021
|
|
6B3O
 
 | | Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion | | Descriptor: | Spike glycoprotein | | Authors: | Walls, A.C, Tortorici, M.A, Snijder, J, Xiong, X, Bosch, B.J, Rey, F.A, Veesler, D. | | Deposit date: | 2017-09-22 | | Release date: | 2017-10-04 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6B40
 
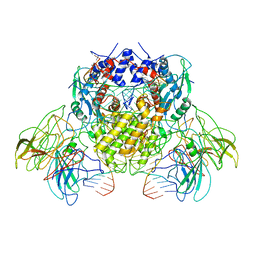 | | BbRAGL-3'TIR synaptic complex with nicked DNA refined with C2 symmetry | | Descriptor: | 31TIR intact strand, 31TIR pre-nicked strand of flanking DNA, 31TIR pre-nicked strand of signal DNA, ... | | Authors: | Zhang, Y, Cheng, T.C, Xiong, Y, Schatz, D.G. | | Deposit date: | 2017-09-25 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Transposon molecular domestication and the evolution of the RAG recombinase.
Nature, 569, 2019
|
|
8F8V
 
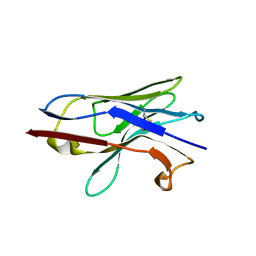 | | Crystal structure of Nb.X0 | | Descriptor: | Nb.X0 | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Andi, B. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
8F8W
 
 | | Crystal structure of Nb.X0 bound to the afucosylated human IgG1 fragment crystal form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb.X0, afucosylated IgG1 fragment | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Oren, D. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
8F8X
 
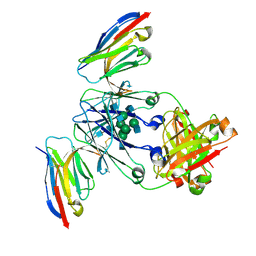 | | Crystal structure of Nb.X0 bound to the afucosylated human IgG1 fragment crystal form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb.X0, Uncharacterized protein DKFZp686C11235 | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Oren, D. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
8TEX
 
 | | Avian Adeno-associated virus - empty capsid | | Descriptor: | Capsid protein | | Authors: | Hsi, J, Mietzsch, M, Chipman, P, Afione, S, Zeher, A, Huang, R, Chiorini, J, McKenna, R. | | Deposit date: | 2023-07-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural and antigenic characterization of the avian adeno-associated virus capsid.
J.Virol., 97, 2023
|
|
8TEY
 
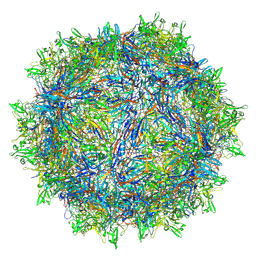 | | Avian Adeno-associated virus - empty capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | Authors: | Hsi, J, Mietzsch, M, Chipman, P, Afione, S, Zeher, A, Huang, R, Chiorini, J, McKenna, R. | | Deposit date: | 2023-07-07 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural and antigenic characterization of the avian adeno-associated virus capsid.
J.Virol., 97, 2023
|
|
6C97
 
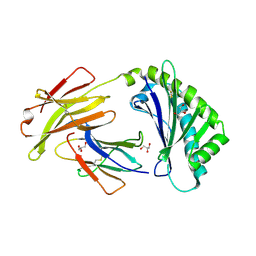 | | Crystal structure of FcRn at pH3 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, IgG receptor FcRn large subunit p51 | | Authors: | Fox III, D, Fairman, J.W. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
7E9G
 
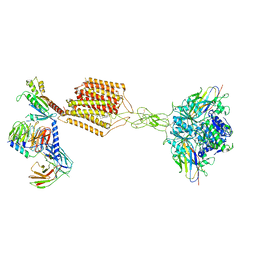 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu2 | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, DN13, ... | | Authors: | Lin, S, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2021-03-04 | | Release date: | 2021-06-23 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
8FMX
 
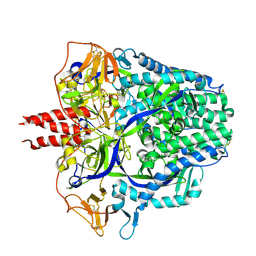 | | Langya virus F glycoprotein ectodomain in prefusion form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Low, Y.S, Isaacs, A, Modhiran, N, Watterson, D. | | Deposit date: | 2022-12-26 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure and antigenicity of divergent Henipavirus fusion glycoproteins.
Nat Commun, 14, 2023
|
|
8FMY
 
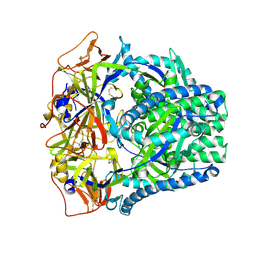 | |
6C99
 
 | | Crystal structure of FcRn bound to UCB-303 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CITRIC ACID, ... | | Authors: | Fox III, D, Abendroth, J, Porter, J, Deboves, H. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
8FEZ
 
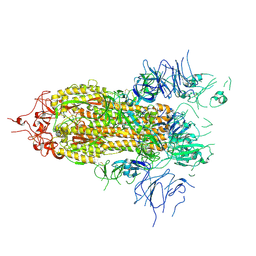 | |
6CJ7
 
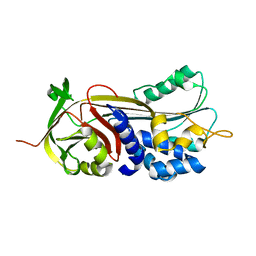 | | Crystal structure of Manduca sexta Serine protease inhibitor (Serpin)-12 | | Descriptor: | Serpin-12 | | Authors: | Gulati, M, Hu, Y, Peng, S, Pathak, P.K, Wang, Y, Deng, J, Jiang, H. | | Deposit date: | 2018-02-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Manduca sexta serpin-12 controls the prophenoloxidase activation system in larval hemolymph.
Insect Biochem. Mol. Biol., 99, 2018
|
|
7F2B
 
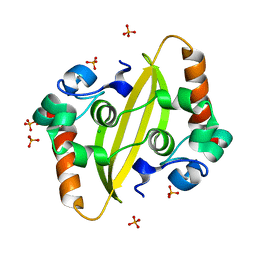 | |
6CRJ
 
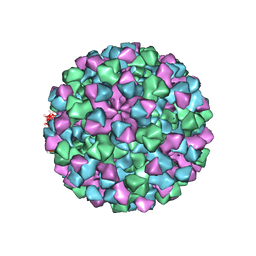 | |
8FTT
 
 | |
7F2E
 
 | | SARS-CoV-2 nucleocapsid protein C-terminal domain (dodecamer) | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Liu, C, Jiang, H. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the SARS-CoV-2 nucleocapsid protein C-terminal domain and development of nucleocapsid-targeting nanobodies.
Febs J., 289, 2022
|
|
6C6Q
 
 | |
6C74
 
 | |
8UUL
 
 | | Prototypic SARS-CoV-2 spike (containing K417) in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
