6I2H
 
 | |
1QFE
 
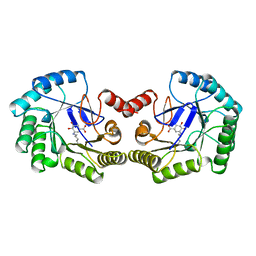 | | THE STRUCTURE OF TYPE I 3-DEHYDROQUINATE DEHYDRATASE FROM SALMONELLA TYPHI | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, PROTEIN (3-DEHYDROQUINATE DEHYDRATASE) | | Authors: | Shrive, A.K, Polikarpov, I, Sawyer, L, Coggins, J.R, Hawkins, A.R. | | Deposit date: | 1999-04-05 | | Release date: | 2000-04-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The two types of 3-dehydroquinase have distinct structures but catalyze the same overall reaction.
Nat.Struct.Biol., 6, 1999
|
|
5X1Y
 
 | |
5X0E
 
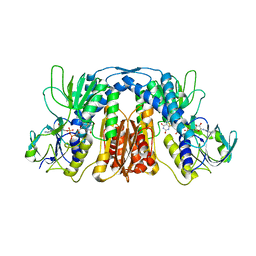
 | | Free serine kinase (E30A mutant) in complex with phosphoserine and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Free serine kinase, MAGNESIUM ION, ... | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2017-01-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Study on the Reaction Mechanism of a Free Serine Kinase Involved in Cysteine Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
1QGJ
 
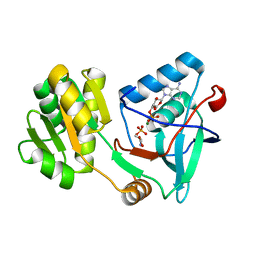
 | | ARABIDOPSIS THALIANA PEROXIDASE N | | Descriptor: | CALCIUM ION, GLUTATHIONE, PEROXIDASE N, ... | | Authors: | Mirza, O, Oestergaard, L, Welinder, K.G, Henriksen, A, Gajhede, M. | | Deposit date: | 1999-04-29 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Arabidopsis thaliana peroxidase N: structure of a novel neutral peroxidase.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5X1C
 
 | |
6I4Z
 
 | | Crystal structure of the disease-causing P453L mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Wilk, P, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
5X20
 
 | | The ternary structure of D-mandelate dehydrogenase with NADH and anilino(oxo)acetate | | Descriptor: | 2-dehydropantoate 2-reductase, 2-oxidanylidene-2-phenylazanyl-ethanoic acid, GLYCEROL, ... | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2017-01-29 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The ternary complex structure of d-mandelate dehydrogenase with NADH and anilino(oxo)acetate.
Biochem. Biophys. Res. Commun., 486, 2017
|
|
5WAH
 
 | | SOLUTION NMR STRUCTURE OF SIGLEC-5 BINDING DOMAIN FROM STREPTOCOCCAL BETA PROTEIN | | Descriptor: | IgA FC receptor | | Authors: | ELETSKY, A, CHEN, C, FONG, J.J, NIZET, V, VARKI, A, PRESTEGARD, J.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | SOLUTION NMR STRUCTURE OF SIGLEC-5 BINDING DOMAIN FROM STREPTOCOCCAL BETA PROTEIN
To Be Published
|
|
1QDR
 
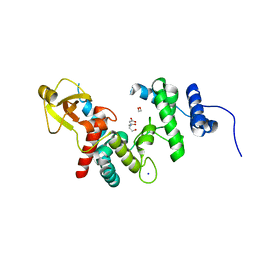 | |
6I62
 
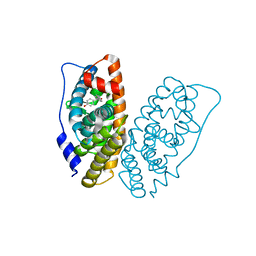 | | Crystal structure of human ERRg LBD in complex with HPTE | | Descriptor: | 4,4'-(2,2,2-trichloroethane-1,1-diyl)diphenol, Estrogen-related receptor gamma | | Authors: | Delfosse, V, Blanc, P, Bourguet, W. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the activation mechanism of human estrogen-related receptor gamma by environmental endocrine disruptors.
Cell.Mol.Life Sci., 76, 2019
|
|
1QE3
 
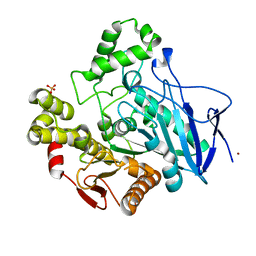 | | PNB ESTERASE | | Descriptor: | PARA-NITROBENZYL ESTERASE, SULFATE ION, ZINC ION | | Authors: | Spiller, B, Gershenson, A, Arnold, F, Stevens, R. | | Deposit date: | 1999-07-12 | | Release date: | 1999-07-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural view of evolutionary divergence.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
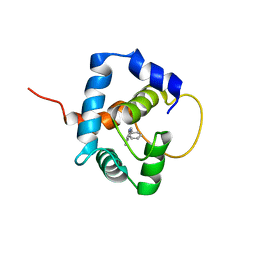
5W88
 
 | |
5WCL
 
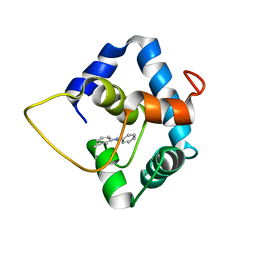 | | NMR structure of the N-domain of troponin C bound to switch region of troponin I and 3-methyldiphenylamine (solvent exposed mode) | | Descriptor: | 3-methyl-N-phenylaniline, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Cai, F, Hwang, P.M, Sykes, B.D. | | Deposit date: | 2017-06-30 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures reveal details of small molecule binding to cardiac troponin.
J. Mol. Cell. Cardiol., 101, 2016
|
|
6I89
 
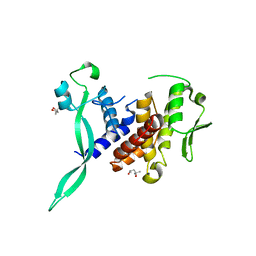 | |
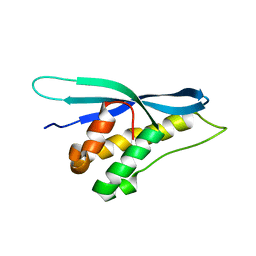
5WOE
 
 | |
1QJT
 
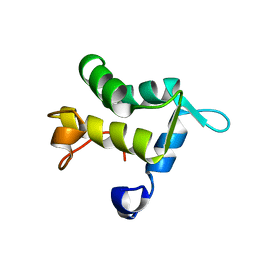 | | SOLUTION STRUCTURE OF THE APO EH1 DOMAIN OF MOUSE EPIDERMAL GROWTH FACTOR RECEPTOR SUBSTRATE 15, EPS15 | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR SUBSTRATE SUBSTRATE 15, EPS15 | | Authors: | Whitehead, B, Tessari, M, Carotenuto, A, van Bergen en Henegouwen, P.M, Vuister, G.W. | | Deposit date: | 1999-07-02 | | Release date: | 2000-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Eh1 Domain of Eps15 is Structurally Classified as a Member of the S100 Subclass of EF-Hand Containing Proteins
Biochemistry, 38, 1999
|
|
5WOY
 
 | |
5WE3
 
 | | Solution NMR structure of PaurTx-3 | | Descriptor: | Beta-theraphotoxin-Ps1a | | Authors: | Agwa, A.J, Schroeder, C.I. | | Deposit date: | 2017-07-06 | | Release date: | 2017-09-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Lengths of the C-Terminus and Interconnecting Loops Impact Stability of Spider-Derived Gating Modifier Toxins.
Toxins (Basel), 9, 2017
|
|
2KUH
 
 | | Halothane binds to druggable sites in calcium-calmodulin: Solution structure of halothane-CaM C-terminal domain | | Descriptor: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, CALCIUM ION, Calmodulin | | Authors: | Juranic, N, Macura, S, Simeonov, M.V, Jones, K.A, Penheiter, A.R, Hock, T.J, Streiff, J.H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Halothane binds to druggable sites in the [Ca2+]4-calmodulin (CaM) complex, but does not inhibit [Ca2+]4-CaM activation of kinase.
J. Serb. Chem. Soc., 78, 2013
|
|
1QG8
 
 | | NATIVE (MAGNESIUM-CONTAINING) SPSA FROM BACILLUS SUBTILIS | | Descriptor: | GLYCEROL, MAGNESIUM ION, PROTEIN (SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA) | | Authors: | Charnock, S.J. | | Deposit date: | 1999-04-21 | | Release date: | 2000-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms.
Biochemistry, 38, 1999
|
|
5W8Y
 
 | |
5WDZ
 
 | |
1QGU
 
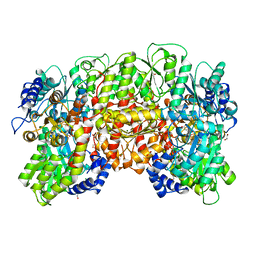 | | NITROGENASE MO-FE PROTEIN FROM KLEBSIELLA PNEUMONIAE, DITHIONITE-REDUCED STATE | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CHLORIDE ION, ... | | Authors: | Mayer, S.M, Lawson, D.M, Gormal, C.A, Roe, S.M, Smith, B.E. | | Deposit date: | 1999-05-06 | | Release date: | 1999-11-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into structure-function relationships in nitrogenase: A 1.6 A resolution X-ray crystallographic study of Klebsiella pneumoniae MoFe-protein.
J.Mol.Biol., 292, 1999
|
|
5WQZ
 
 | |
