7PMO
 
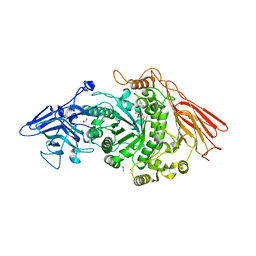 | | Ruminococcus gnavus ATC29149 endo-beta-1,4-galactosidase (RgGH98) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Owen, C.D, Wu, H, Crost, E.H, van Bakel, W, Gascuena, A.M, Latousakis, D, Hicks, T, Walpole, S, Urbanowicz, P.A, Ndeh, D, Monaco, S, Salom, L.S, Griffiths, R, Colvile, A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2021-09-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging: Implication for niche colonisation in the gastrointestinal tract.
Plos Biol., 19, 2021
|
|
7PU8
 
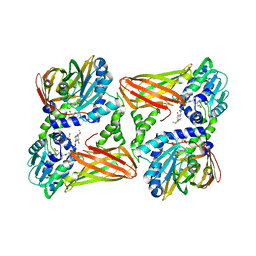 | | CARM1 in complex with EML980 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[N-[2-[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-09-28 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7PUC
 
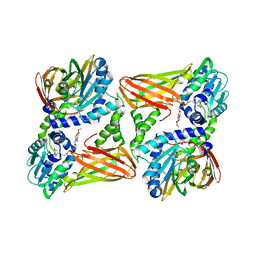 | | CARM1 in complex with EML981 | | Descriptor: | 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, methyl 6-[4-[[~{N}-[(~{E})-3-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]prop-2-enyl]carbamimidoyl]amino]butylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Marechal, N, Cura, V, Bonnefond, L, Troffer-Charlier, N, Cavarelli, J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
6PGE
 
 | |
7PW1
 
 | | Crystal structure of ancestral haloalkane dehalogenase AncLinB-DmbA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Mazur, A, Grinkevich, P, Prudnikova, T. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of the Ancestral Haloalkane Dehalogenase AncLinB-DmbA.
Int J Mol Sci, 22, 2021
|
|
6P58
 
 | | Dark and Steady State-Illuminated Crystal Structure of Cyanobacteriochrome Receptor PixJ at 150K | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Buirgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-05-29 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4X1R
 
 | | The crystal structure of mupain-1-12 in complex with murinised human uPA at pH7.4 | | Descriptor: | 1-phenylguanidine, Urokinase-type plasminogen activator, mupain-1-12 | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
4XPN
 
 | | Crystal Structure of Protein Phosphate 1 complexed with PP1 binding domain of GADD34 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2015-01-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Structural and Functional Analysis of the GADD34:PP1 eIF2 alpha Phosphatase.
Cell Rep, 11, 2015
|
|
6OA1
 
 | | Structure of human PARG complexed with JA2120 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
7PWR
 
 | | PARP15 catalytic domain in complex with OUL254 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-(cyclopentylmethoxy)phthalazine-1,4-dione, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-10-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
7PWL
 
 | | PARP15 catalytic domain in complex with OUL218 | | Descriptor: | 4-(1,3-thiazol-5-ylmethoxy)benzamide, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Murthy, S, Lehtio, L. | | Deposit date: | 2021-10-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
7PWS
 
 | | PARP15 catalytic domain in complex with OUL255 | | Descriptor: | 6-(cyclobutylmethoxy)phthalazine-1,4-dione, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-10-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
7PWW
 
 | | PARP15 catalytic domain in complex with OUL224 | | Descriptor: | 6-[(3-bromophenyl)methoxy]phthalazine-1,4-dione, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Murthy, S, Nizi, M.G, Lehtio, L. | | Deposit date: | 2021-10-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
7PWU
 
 | | PARP15 catalytic domain in complex with OUL256 | | Descriptor: | 6-(cyclopropylmethoxy)phthalazine-1,4-dione, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-10-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
4XUC
 
 | | Synthesis and evaluation of heterocyclic catechol mimics as inhibitors of catechol-O-methyltransferase (COMT): Structure with Cmpd18 (1-(biphenyl-3-yl)-3-hydroxypyridin-4(1H)-one) | | Descriptor: | 1-(biphenyl-3-yl)-3-hydroxypyridin-4(1H)-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Catechol O-methyltransferase, ... | | Authors: | Allison, T, Wolkenberg, S, Sanders, J.M, Soisson, S.M. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT).
Acs Med.Chem.Lett., 6, 2015
|
|
4XPP
 
 | | Crystal structure of Pedobacter saltans GH31 alpha-galactosidase complexed with D-galactose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-glucosidase, beta-D-galactopyranose | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
9S5G
 
 | | Temporizin-1 NMR analysis | | Descriptor: | Temporizin-1 | | Authors: | Falcigno, L, D'Auria, G, Galdiero, S, Falanga, A, Bellavita, R. | | Deposit date: | 2025-07-29 | | Release date: | 2025-09-17 | | Last modified: | 2025-10-08 | | Method: | SOLUTION NMR | | Cite: | Temporizin-1 Meets the Membranes: Probing Membrane Inser-Tion and Disruption Mechanisms.
Antibiotics, 14, 2025
|
|
6NWK
 
 | |
4W9Q
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
9U7V
 
 | | The X-ray Crystallographic Structure of oligo-1,6-glucosidase from Paenibacillus sp. STB16 | | Descriptor: | oligo-1,6-glucosidase | | Authors: | Li, Z.F, Tian, Y.X, Zhang, J.S, Ban, X.F. | | Deposit date: | 2025-03-25 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | The X-ray Crystallographic Structure of oligo-1,6-glucosidase from Paenibacillus sp. STB16
To Be Published
|
|
6NXG
 
 | | Crystal structure of glycylpeptide N-tetradecanoyltransferase from Plasmodium vivax in complex with inhibitor 303a | | Descriptor: | 1,2-ETHANEDIOL, 5-(4-chlorophenyl)-3-({[3-(morpholine-4-carbonyl)phenyl]amino}methyl)pyridin-2(1H)-one, BETA-MERCAPTOETHANOL, ... | | Authors: | Staker, B.L, Mayclin, S, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-02-08 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of Selective Inhibitors ofPlasmodiumN-Myristoyltransferase by High-Throughput Screening.
J.Med.Chem., 63, 2020
|
|
4UTR
 
 | | Crystal structure of zebrafish Sirtuin 5 in complex with glutarylated CPS1-peptide | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GLUTARIC ACID, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6O0N
 
 | | M.tb MenD with Inhibitor | | Descriptor: | 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Johnston, J.M, Ho, N.A.T, Bashiri, G, Bulloch, E.M, Nigon, L.V, Jirgis, E.M.N, Baker, E.N. | | Deposit date: | 2019-02-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
7P07
 
 | | Structure of the maltase BaAG2 from Blastobotrys adeninivorans in complex with glucose | | Descriptor: | 1-methylpyrrolidin-2-one, BaAG2, CALCIUM ION, ... | | Authors: | Ernits, K, Visnapuu, T, Persson, K. | | Deposit date: | 2021-06-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Insight into a Yeast Maltase-The Ba AG2 from Blastobotrys adeninivorans with Transglycosylating Activity.
J Fungi (Basel), 7, 2021
|
|
4W79
 
 | | Crystal Structure of Human Protein N-terminal Glutamine Amidohydrolase | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Protein N-terminal glutamine amidohydrolase, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human protein N-terminal glutamine amidohydrolase, an initial component of the N-end rule pathway.
Plos One, 9, 2014
|
|
