5HJM
 
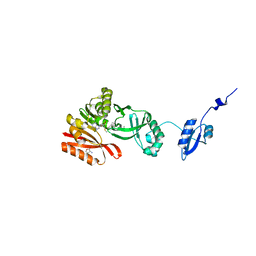 | | Crystal Structure of Pyrococcus abyssi Trm5a complexed with MTA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ACETATE ION, HOMOSERINE LACTONE, ... | | Authors: | Xie, W, Wang, C, Jia, Q. | | Deposit date: | 2016-01-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Crystal structures of the bifunctional tRNA methyltransferase Trm5a
Sci Rep, 6, 2016
|
|
1CFE
 
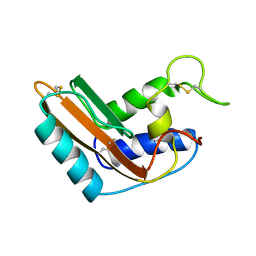 | | P14A, NMR, 20 STRUCTURES | | Descriptor: | PATHOGENESIS-RELATED PROTEIN P14A | | Authors: | Fernandez, C, Szyperski, T, Bruyere, T, Ramage, P, Mosinger, E, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the pathogenesis-related protein P14a.
J.Mol.Biol., 266, 1997
|
|
1F7Z
 
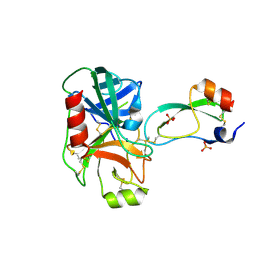 | | RAT TRYPSINOGEN K15A COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR | | Descriptor: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION, ... | | Authors: | Pasternak, A, White, A, Jeffery, C.J, Medina, N, Cahoon, M, Ringe, D, Hedstrom, L. | | Deposit date: | 2000-06-28 | | Release date: | 2001-07-04 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The energetic cost of induced fit catalysis: Crystal structures of trypsinogen mutants with enhanced activity and inhibitor affinity.
Protein Sci., 10, 2001
|
|
6W14
 
 | |
5HJI
 
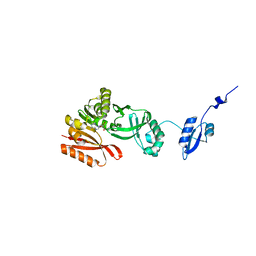 | |
1OY5
 
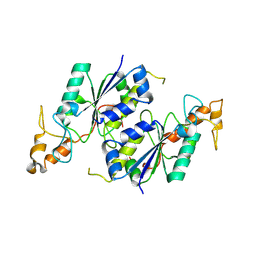 | | Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus | | Descriptor: | tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Liu, J, Wang, W, Shin, D.H, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-04-03 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus at 2.6 A resolution: a novel methyltransferase fold.
Proteins, 53, 2003
|
|
1DYW
 
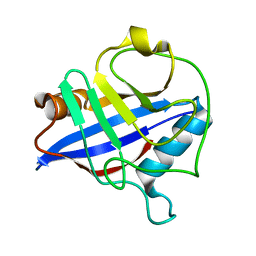 | | Biochemical and structural characterization of a divergent loop cyclophilin from Caenorhabditis elegans | | Descriptor: | CYCLOPHILIN 3 | | Authors: | Dornan, J, Page, A.P, Taylor, P, Wu, S.Y, Winter, A.D, Husi, H, Walkinshaw, M.D. | | Deposit date: | 2000-02-10 | | Release date: | 2000-06-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and Structural Characterization of a Divergent Loop Cyclophilin from Caenorhabditis Elegans
J.Biol.Chem., 274, 1999
|
|
1H61
 
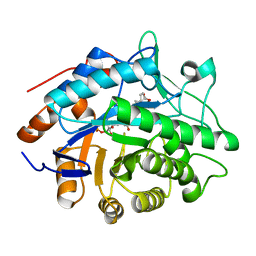 | | Structure of Pentaerythritol Tetranitrate Reductase in complex with prednisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1FY8
 
 | | CRYSTAL STRUCTURE OF THE DELTAILE16VAL17 RAT ANIONIC TRYPSINOGEN-BPTI COMPLEX | | Descriptor: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION, ... | | Authors: | Pasternak, A, White, A, Jeffery, C.J, Ringe, D, Hedstrom, L. | | Deposit date: | 2000-09-28 | | Release date: | 2000-11-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The energetic cost of induced fit catalysis: Crystal structures of trypsinogen mutants with enhanced activity and inhibitor affinity.
Protein Sci., 10, 2001
|
|
1GWJ
 
 | | Morphinone reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, MORPHINONE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2002-03-18 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Bacterial Morphinone Reductase and Properties of the C191A Mutant Enzyme.
J.Biol.Chem., 277, 2002
|
|
1EMX
 
 | | SOLUTION STRUCTURE OF HPTX2, A TOXIN FROM HETEROPODA VENATORIA SPIDER VENOM THAT BLOCKS KV4.2 POTASSIUM CHANNEL | | Descriptor: | HETEROPODATOXIN 2 | | Authors: | Bernard, C, Legros, C, Ferrat, G, Bishoff, U, Marquardt, A, Pongs, O, Darbon, H. | | Deposit date: | 2000-03-20 | | Release date: | 2001-01-24 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hpTX2, a toxin from Heteropoda venatoria spider that blocks Kv4.2 potassium channel.
Protein Sci., 9, 2000
|
|
1H63
 
 | | Structure of the reduced Pentaerythritol Tetranitrate Reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
1H60
 
 | |
1H62
 
 | | Structure of Pentaerythritol tetranitrate reductase in complex with 1,4-androstadien-3,17-dione | | Descriptor: | ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
4BFQ
 
 | | Assembly of a triple pi-stack of ligands in the binding site of Aplysia californica acetylcholine binding protein (AChBP) | | Descriptor: | 4,6-dimethyl-N'-(3-pyridin-2-ylisoquinolin-1-yl)pyrimidine-2-carboximidamide, GLYCEROL, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Stornaiuolo, M, De Kloe, G.E, Rucktooa, P, Fish, A, van Elk, R, Edink, E.S, Bertrand, D, Smit, A.B, de Esch, I.J.P, Sixma, T.K. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Assembly of a Pi-Pi Stack of Ligands in the Binding Site of an Acetylcholine Binding Protein
Nat.Commun., 4, 2013
|
|
1K5W
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE SYNAPTOTAGMIN 1 C2B-DOMAIN: SYNAPTOTAGMIN 1 AS A PHOSPHOLIPID BINDING MACHINE | | Descriptor: | CALCIUM ION, Synaptotagmin I | | Authors: | Fernandez, I, Arac, D, Ubach, J, Gerber, S.H, Shin, O, Gao, Y, Anderson, R.G.W, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-10-12 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine.
Neuron, 32, 2001
|
|
1XFQ
 
 | | structure of the blue shifted intermediate state of the photoactive yellow protein lacking the N-terminal part | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Bernard, C, Houben, K, Derix, N.M, Marks, D, van der Horst, M.A, Hellingwerf, K.J, Boelens, R, Kaptein, R, van Nuland, N.A. | | Deposit date: | 2004-09-15 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a transient photoreceptor intermediate: delta25 photoactive yellow protein
STRUCTURE, 13, 2005
|
|
1ZHG
 
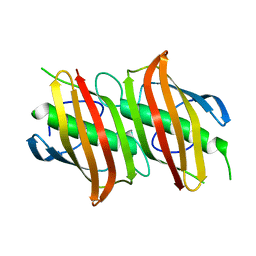 | | Crystal structure of Beta-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Plasmodium falciparum | | Descriptor: | beta hydroxyacyl-acyl carrier protein dehydratase | | Authors: | Swarnamukhi, P.L, Sharma, S.K, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2005-04-25 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of dimeric FabZ of Plasmodium falciparum reveals conformational switching to active hexamers by peptide flips
Febs Lett., 580, 2006
|
|
4CSD
 
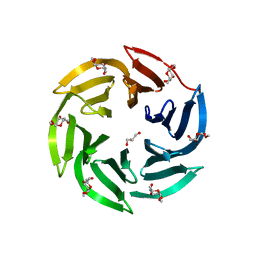 | | Structure of Monomeric Ralstonia solanacearum lectin | | Descriptor: | FUCOSE-BINDING LECTIN PROTEIN, GLYCEROL, methyl alpha-L-fucopyranoside | | Authors: | Arnaud, J, Trundle, K, Claudinon, J, Audfray, A, Varrot, A, Romer, W, Imberty, A. | | Deposit date: | 2014-03-06 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Membrane Deformation by Neolectins with Engineered Glycolipid Binding Sites.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1D6N
 
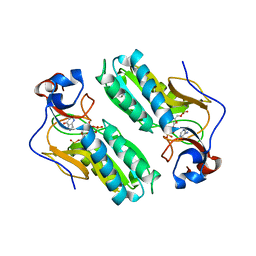 | | TERNARY COMPLEX STRUCTURE OF HUMAN HGPRTASE, PRPP, MG2+, AND THE INHIBITOR HPP REVEALS THE INVOLVEMENT OF THE FLEXIBLE LOOP IN SUBSTRATE BINDING | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 3H-PYRAZOLO[4,3-D]PYRIMIDIN-7-OL, MAGNESIUM ION, ... | | Authors: | Balendiran, G.K. | | Deposit date: | 1999-10-14 | | Release date: | 1999-12-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ternary complex structure of human HGPRTase, PRPP, Mg2+, and the inhibitor HPP reveals the involvement of the flexible loop in substrate binding.
Protein Sci., 8, 1999
|
|
1KTX
 
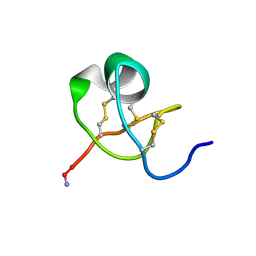 | | KALIOTOXIN (1-37) SHOWS STRUCTURAL DIFFERENCES WITH RELATED POTASSIUM CHANNEL BLOCKERS | | Descriptor: | KALIOTOXIN | | Authors: | Fernandez, I, Romi, R, Szendefi, S, Martin-Eauclaire, M.-F, Rochat, H, Van Rietschtoten, J, Pons, M, Giralt, E. | | Deposit date: | 1994-06-02 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Kaliotoxin (1-37) shows structural differences with related potassium channel blockers.
Biochemistry, 33, 1994
|
|
1XFN
 
 | | NMR structure of the ground state of the photoactive yellow protein lacking the N-terminal part | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Bernard, C, Houben, K, Derix, N.M, Marks, D, van der Horst, M.A, Hellingwerf, K.J, Boelens, R, Kaptein, R, van Nuland, N.A. | | Deposit date: | 2004-09-15 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a transient photoreceptor intermediate: delta25 photoactive yellow protein
STRUCTURE, 13, 2005
|
|
1I26
 
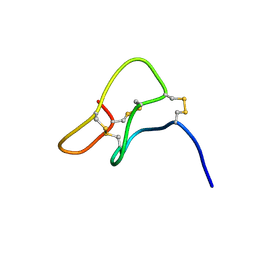 | | SOLUTION STRUCTURE OF PTU-1, A TOXIN FROM THE ASSASSIN BUGS PEIRATES TURPIS THAT BLOCKS THE VOLTAGE SENSITIVE CALCIUM CHANNEL N-TYPE | | Descriptor: | PTU-1 | | Authors: | Bernard, C, Corzo, G, Mosbah, A, Nakajima, T, Darbon, H. | | Deposit date: | 2001-02-06 | | Release date: | 2001-11-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ptu1, a toxin from the assassin bug Peirates turpis that blocks the voltage-sensitive calcium channel N-type.
Biochemistry, 40, 2001
|
|
1LMR
 
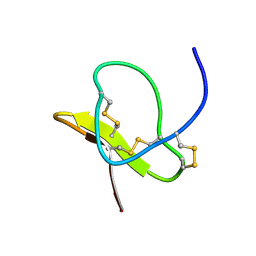 | | Solution of ADO1, a Toxin from the Assassin Bugs Agriosphodrus dohrni that Blocks the Voltage Sensitive Calcium Channel L-type | | Descriptor: | TOXIN ADO1 | | Authors: | Bernard, C, Corzo, G, Adachi-Akahane, S, Foures, G, Kanemaru, K, Furukawa, Y, Nakajima, T, Darbon, H. | | Deposit date: | 2002-05-02 | | Release date: | 2003-08-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ADO1, a toxin extracted from the saliva of the assassin bug, Agriosphodrus dohrni
Proteins: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
1XXG
 
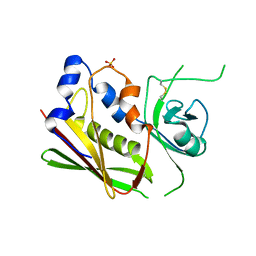 | |
