2WYP
 
 | | Crystal structure of sialic acid binding protein | | Descriptor: | SIALIC ACID-BINDING PERIPLASMIC PROTEIN SIAP, deamino-beta-neuraminic acid | | Authors: | Fischer, M, Hubbard, R.E. | | Deposit date: | 2009-11-18 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water networks can determine the affinity of ligand binding to proteins.
J.Am.Chem.Soc., 2019
|
|
6YGG
 
 | | NADase from Aspergillus fumigatus complexed with a substrate anologue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, AfNADase, ... | | Authors: | Stromland, O, Ziegler, M, Kallio, J.P. | | Deposit date: | 2020-03-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of fungal surface NADases predominantly present in pathogenic species.
Nat Commun, 12, 2021
|
|
3TGT
 
 | | Crystal structure of unliganded HIV-1 clade A/E strain 93TH057 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 clade A/E 93TH057 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6YGF
 
 | | NADase from Aspergillus fumigatus with trapped reaction products | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, AfNADase, ... | | Authors: | Stromland, O, Ziegler, M, Kallio, J.P. | | Deposit date: | 2020-03-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of fungal surface NADases predominantly present in pathogenic species.
Nat Commun, 12, 2021
|
|
6YGE
 
 | | NADase from Aspergillus fumigatus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, AfNADase, ... | | Authors: | Stromland, O, Ziegler, M, Kallio, J.P. | | Deposit date: | 2020-03-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of fungal surface NADases predominantly present in pathogenic species.
Nat Commun, 12, 2021
|
|
3TGQ
 
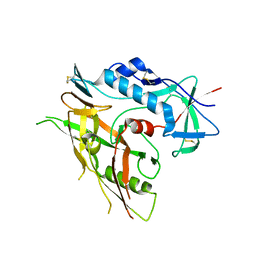 | | Crystal structure of unliganded HIV-1 clade B strain YU2 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 YU2 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1JNQ
 
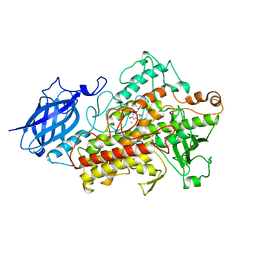 | | LIPOXYGENASE-3 (SOYBEAN) COMPLEX WITH EPIGALLOCATHECHIN (EGC) | | Descriptor: | 2-(3,4,5-TRIHYDROXY-PHENYL)-CHROMAN-3,5,7-TRIOL, FE (II) ION, lipoxygenase-3 | | Authors: | Zhou, K, Skrzypczak-Jankun, E, Jankun, J. | | Deposit date: | 2001-07-24 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of lipoxygenase by (-)-epigallocatechin gallate: X-ray analysis at 2.1 A reveals degradation of EGCG
and shows soybean LOX-3 complex with EGC instead.
INT.J.MOL.MED., 12, 2003
|
|
4C1Y
 
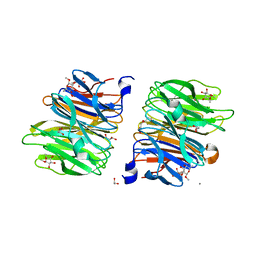 | | Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) in complex with b-methylfucoside | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, FUCOSE-SPECIFIC LECTIN FLEA, ... | | Authors: | Houser, J, Komarek, J, Kostlanova, N, Lahmann, M, Cioci, G, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2013-08-14 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural Insights Into Aspergillus Fumigatus Lectin Specificity: Afl Binding Sites are Functionally Non-Equivalent.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2XA5
 
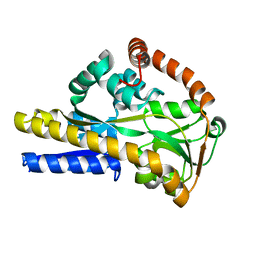 | |
3TGR
 
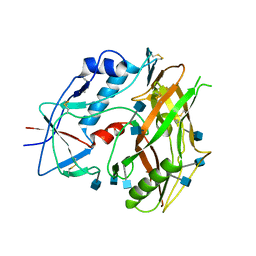 | | Crystal structure of unliganded HIV-1 clade C strain C1086 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6X1N
 
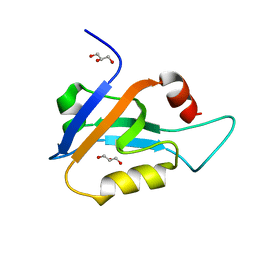 | |
2XCY
 
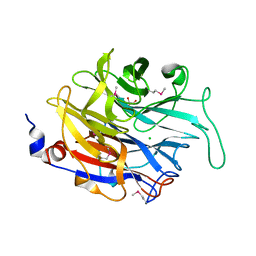 | | Crystal structure of Aspergillus fumigatus sialidase | | Descriptor: | CHLORIDE ION, EXTRACELLULAR SIALIDASE/NEURAMINIDASE, PUTATIVE, ... | | Authors: | Telford, J.C, Yeung, J, Xu, G, Bennet, A, Moore, M.M, Taylor, G.L. | | Deposit date: | 2010-04-27 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Aspergillus Fumigatus Sialidase is a Kdnase: Structural and Mechanistic Insights.
J.Biol.Chem., 286, 2011
|
|
2DSX
 
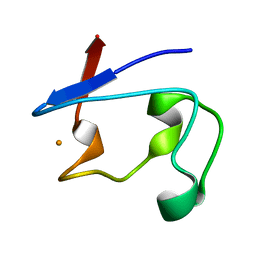 | | Crystal structure of rubredoxin from Desulfovibrio gigas to ultra-high 0.68 A resolution | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Chen, C.-J, Lin, Y.-H, Huang, Y.-C, Liu, M.-Y. | | Deposit date: | 2006-07-07 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.68 Å) | | Cite: | Crystal structure of rubredoxin from Desulfovibrio gigas to ultra-high 0.68A resolution
Biochem.Biophys.Res.Commun., 349, 2006
|
|
1QN1
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERRICYTOCHROME C3, NMR, 15 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Brennan, L, Messias, A.C, Legall, J, Turner, D.L, Xavier, A.V. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochromes C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
2PHN
 
 | | Crystal structure of an amide bond forming F420-gamma glutamyl ligase from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, F420-0:gamma-glutamyl ligase, ... | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-11 | | Release date: | 2007-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of an Amide Bond Forming F(420):gammagamma-glutamyl Ligase from Archaeoglobus Fulgidus - A Member of a New Family of Non-ribosomal Peptide Synthases.
J.Mol.Biol., 372, 2007
|
|
1QN0
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Teodoro, M.L, Brennan, L, Legall, J, Santos, H, Xavier, A.V, Turner, D.L. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochrome C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
1GYO
 
 | | Crystal structure of the di-tetraheme cytochrome c3 from Desulfovibrio gigas at 1.2 Angstrom resolution | | Descriptor: | CYTOCHROME C3, A DIMERIC CLASS III C-TYPE CYTOCHROME, GLYCEROL, ... | | Authors: | Aragao, D, Frazao, C, Sieker, L, Sheldrick, G.M, Legall, J, Carrondo, M.A. | | Deposit date: | 2002-04-29 | | Release date: | 2002-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Dimeric Cytochrome C3 from Desulfovibrio Gigas at 1.2 A Resolution
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6APW
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | 4-[(4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)methyl]benzoic acid, Bifunctional ligase/repressor BirA | | Authors: | Cini, D.A, Wilce, M.C.J. | | Deposit date: | 2017-08-18 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.614 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
6AQQ
 
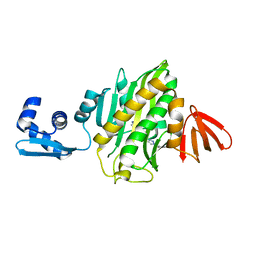 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | (3aS,4S,6aR)-4-(5-{1-[(3-fluorophenyl)methyl]-1H-1,2,3-triazol-4-yl}pentyl)tetrahydro-1H-thieno[3,4-d]imidazol-2(3H)-one, Bifunctional ligase/repressor BirA | | Authors: | Cini, D.A, Wilce, M.C.J. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
1FC4
 
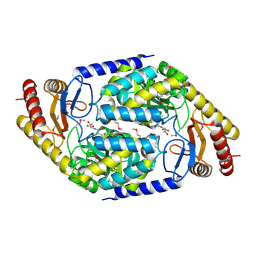 | | 2-AMINO-3-KETOBUTYRATE COA LIGASE | | Descriptor: | 2-AMINO-3-KETOBUTYRATE CONENZYME A LIGASE, 2-AMINO-3-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schmidt, A, Matte, A, Li, Y, Sivaraman, J, Larocque, R, Schrag, J.D, Smith, C, Sauve, V, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2000-07-17 | | Release date: | 2001-05-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of 2-amino-3-ketobutyrate CoA ligase from Escherichia coli complexed with a PLP-substrate intermediate: inferred reaction mechanism.
Biochemistry, 40, 2001
|
|
4PZP
 
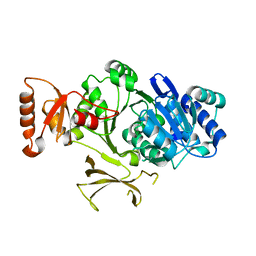 | |
5LP8
 
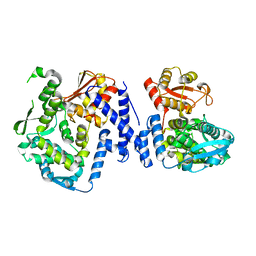 | |
2KR1
 
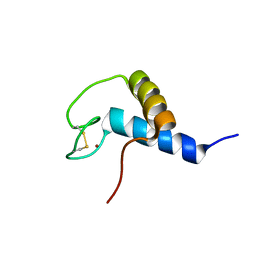 | | Solution NMR structure of zinc binding N-terminal domain of ubiquitin-protein ligase E3A from Homo Sapiens. Northeast Structural Genomics Consortium (NESG) target HR3662 | | Descriptor: | Ubiquitin protein ligase E3A, ZINC ION | | Authors: | Lemak, A, Yee, A, Fares, C, Semesi, A, Xiao, R, Montelione, G, Dhe-Paganon, S, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-27 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Zn-binding AZUL domain of human ubiquitin protein ligase Ube3A.
J.Biomol.Nmr, 51, 2011
|
|
3TSY
 
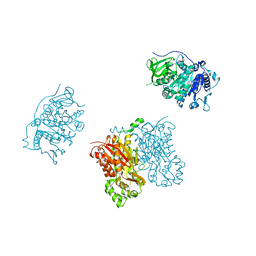 | | 4-Coumaroyl-CoA Ligase::Stilbene Synthase fusion protein | | Descriptor: | Fusion Protein 4-coumarate--CoA ligase 1, Resveratrol synthase | | Authors: | Yi, H, Jez, J.M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Kinetic Analysis of the Unnatural Fusion Protein 4-Coumaroyl-CoA Ligase::Stilbene Synthase.
J.Am.Chem.Soc., 133, 2011
|
|
8UYO
 
 | |
