5Z8F
 
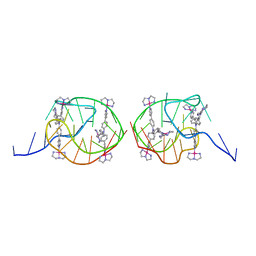 | | Solution structure for the unique dimeric 4:2 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | Descriptor: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | Authors: | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | Deposit date: | 2018-01-31 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
8GXT
 
 | |
8HJD
 
 | |
8HJC
 
 | |
7OVC
 
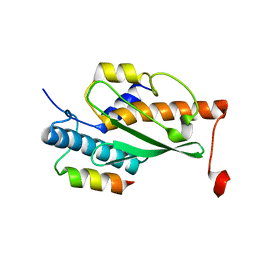 | | Structure of the human UFC1 protein in complex with the UBA5 C-terminal UFC1-binding motif. | | Descriptor: | Ubiquitin-fold modifier-conjugating enzyme 1, Ubiquitin-like modifier-activating enzyme 5 | | Authors: | Wesch, W, Loehr, F, Rogova, N, Doetsch, V, Rogov, V.V. | | Deposit date: | 2021-06-14 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Concerted Action of UBA5 C-Terminal Unstructured Regions Is Important for Transfer of Activated UFM1 to UFC1.
Int J Mol Sci, 22, 2021
|
|
7OXF
 
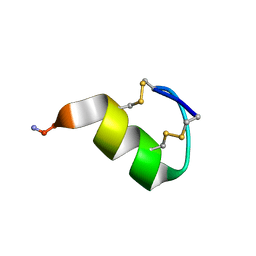 | |
7PDU
 
 | |
8IEX
 
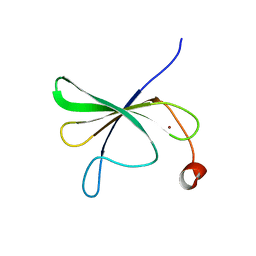 | | Solution structure of AtWRKY11-DBD | | Descriptor: | Probable WRKY transcription factor 11, ZINC ION | | Authors: | Dong, X, Hu, Y.F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of AtWRKY11-DBD
To Be Published
|
|
6AWK
 
 | |
8IC9
 
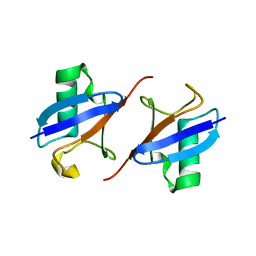 | | Lys48-linked K48C-diubiquitin | | Descriptor: | Polyubiquitin-B, Ubiquitin | | Authors: | Hiranyakorn, M, Yagi-Utsumi, M, Yanaka, S, Ohtsuka, N, Momiyama, N, Satoh, T, Kato, K. | | Deposit date: | 2023-02-11 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mutational and Environmental Effects on the Dynamic Conformational Distributions of Lys48-Linked Ubiquitin Chains.
Int J Mol Sci, 24, 2023
|
|
5TGG
 
 | |
8J4I
 
 | | Unveiling the Role of Human Prohibitin 2 as a Mitochondrial Calcium Channel in Parkinson's Disease | | Descriptor: | Prohibitin-2 | | Authors: | Li, Y.Y, Fang, Y, Gao, Y.X, Ding, W, Yang, J, Liu, Y, Shen, B, Wang, J.F, Zhou, S. | | Deposit date: | 2023-04-20 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Human Prohibitin 2 is a Mitochondrial Ca2+-Selective Channel
To Be Published
|
|
6CKF
 
 | | Structure of a new ShKT peptide from the sea anemone Oulactis sp: OspTx2a-p2 | | Descriptor: | OspTx2a-p2 | | Authors: | Sunanda, P, Krishnarjuna, B, Norton, R.S. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification, chemical synthesis, structure, and function of a new KV1 channel blocking peptide from Oulactis sp.
Peptide Science, 110, 2018
|
|
6CKD
 
 | | Structure of a new ShKT peptide from the sea anemone Oulactis sp: OspTx2a-p1 | | Descriptor: | OspTx2a-p1 | | Authors: | Sunanda, P, Krishnarjuna, B, Norton, R.S. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification, chemical synthesis, structure, and function of a new KV1 channel blocking peptide from Oulactis sp.
Peptide Science, 110, 2018
|
|
2XWR
 
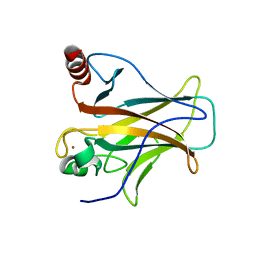 | |
4CA9
 
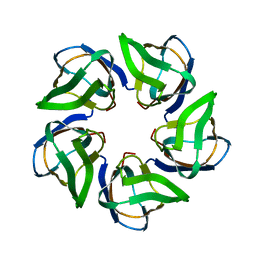 | | Structure of the Nucleoplasmin-like N-terminal domain of Drosophila FKBP39 | | Descriptor: | 39 KDA FK506-BINDING NUCLEAR PROTEIN | | Authors: | Artero, J, Forsyth, T, Callow, P, Watson, A.A, Zhang, W, Laue, E.D, Edlich-Muth, C, Przewloka, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Pentameric Nucleoplasmin Fold is Present in Drosophila Fkbp39 and a Large Number of Chromatin-Related Proteins.
J.Mol.Biol., 427, 2015
|
|
7Q4L
 
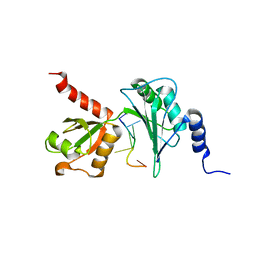 | |
7DME
 
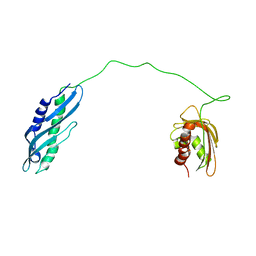 | | Solution structure of human Aha1 | | Descriptor: | Activator of 90 kDa heat shock protein ATPase homolog 1 | | Authors: | Hu, H, Zhou, C, Zhang, N. | | Deposit date: | 2020-12-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Aha1 Exhibits Distinctive Dynamics Behavior and Chaperone-Like Activity.
Molecules, 26, 2021
|
|
7DMD
 
 | |
7EXY
 
 | |
8T0G
 
 | |
8T0H
 
 | |
8T0I
 
 | |
1EIK
 
 | | Solution Structure of RNA Polymerase Subunit RPB5 from Methanobacterium Thermoautotrophicum | | Descriptor: | RNA POLYMERASE SUBUNIT RPB5 | | Authors: | Yee, A, Booth, V, Dharamsi, A, Engel, A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-25 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA polymerase subunit RPB5 from Methanobacterium thermoautotrophicum.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5N7Y
 
 | | Solution structure of B. subtilis Sigma G inhibitor CsfB | | Descriptor: | Anti-sigma-G factor Gin, ZINC ION | | Authors: | Martinez-Lumbreras, S, Alfano, C, Atkinson, A, Isaacson, R.L. | | Deposit date: | 2017-02-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Insights into Bacillus subtilis Sigma Factor Inhibitor, CsfB.
Structure, 26, 2018
|
|
