2WNZ
 
 | | Structure of the E192N mutant of E. coli N-acetylneuraminic acid lyase in complex with pyruvate in space group P21 crystal form I | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 2-ETHOXYETHANOL, LACTIC ACID, ... | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WNQ
 
 | | Structure of the E192N mutant of E. coli N-acetylneuraminic acid lyase in space group P21 | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-17 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
6IDJ
 
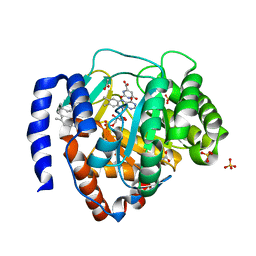 | | Crystal structure of human DHODH in complex with ferulenol | | Descriptor: | 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Shiba, T, Inaoka, D.K, Sato, D, Hartuti, E.D, Amalia, E, Nagahama, M, Yoshioka, Y, Matsubayashi, M, Balogun, E.O, Tsuji, N, Kita, K, Harada, S. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biochemical Features of Eimeria tenella Dihydroorotate Dehydrogenase, a Potential Drug Target.
Genes (Basel), 11, 2020
|
|
9CCS
 
 | |
9CCU
 
 | |
6A4R
 
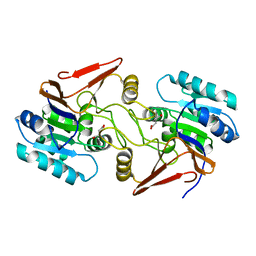 | | Crystal structure of aspartate bound peptidase E from Salmonella enterica | | Descriptor: | ASPARTIC ACID, Peptidase E | | Authors: | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
1LWC
 
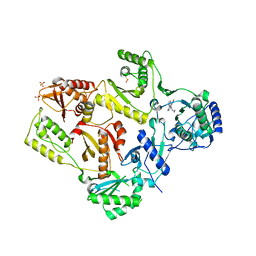 | | CRYSTAL STRUCTURE OF M184V MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
2WO5
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form I | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WNN
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in complex with pyruvate in space group P21 | | Descriptor: | N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL, SODIUM ION | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-13 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
6HZU
 
 | | HUMAN JAK1 IN COMPLEX WITH LASW1393 | | Descriptor: | 2-[4-[8-oxidanylidene-2-[(~{E})-(2-oxidanylidenepyridin-3-ylidene)amino]-7~{H}-purin-9-yl]cyclohexyl]ethanenitrile, Tyrosine-protein kinase JAK1 | | Authors: | Lozoya, E, Segarra, V, Bach, J, Jestel, A, Lammens, A, Blaesse, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of 2-Imidazopyridine and 2-Aminopyridone Purinones as Potent Pan-Janus Kinase (JAK) Inhibitors for the Inhaled Treatment of Respiratory Diseases.
J.Med.Chem., 62, 2019
|
|
5FMG
 
 | | Structure and function based design of Plasmodium-selective proteasome inhibitors | | Descriptor: | (2S)-N-[(E,2S)-1-(1H-indol-3-yl)-4-methylsulfonyl-but-3-en-2-yl]-2-[[(2S)-3-(1H-indol-3-yl)-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]-4-methyl-pentanamide, BETA3 PROTEASOME SUBUNIT, PUTATIVE, ... | | Authors: | Li, H, O'Donoghue, A.J, van der Linden, W.A, Xie, S.C, Yoo, E, Foe, I.T, Tilley, L, Craik, C.S, da Fonseca, P.C.A, Bogyo, M. | | Deposit date: | 2015-11-04 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and Function Based Design of Plasmodium-Selective Proteasome Inhibitors
Nature, 530, 2016
|
|
1W98
 
 | | The structural basis of CDK2 activation by cyclin E | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, G1/S-SPECIFIC CYCLIN E1 | | Authors: | Lowe, E.D, Honda, R, Dubinina, E, Skamnaki, V, Cook, A, Johnson, L.N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of Cyclin E1/Cdk2: Implications for Cdk2 Activation and Cdk2-Independent Roles
Embo J., 24, 2005
|
|
2WPB
 
 | | Crystal structure of the E192N mutant of E. Coli N-acetylneuraminic acid lyase in complex with pyruvate and the inhibitor (2R,3R)-2,3,4- trihydroxy-N,N-dipropylbutanamide in space group P21 crystal form I | | Descriptor: | (2R,3R)-2,3,4-TRIHYDROXY-N,N-DIPROPYLBUTANAMIDE, N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-08-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
6AJE
 
 | | Crystal structure of DHODH in complex with ferulenol from Eimeria tenella | | Descriptor: | 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Shiba, T, Inaoka, D.K, Sato, D, Hartuti, E.D, Amalia, E, Nagahama, M, Yoshioka, Y, Matsubayashi, M, Balogun, E.O, Tsuji, N, Kita, K, Harada, S. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural and Biochemical Features of Eimeria tenella Dihydroorotate Dehydrogenase, a Potential Drug Target.
Genes (Basel), 11, 2020
|
|
9CCT
 
 | |
9CCW
 
 | |
7ZLV
 
 | | Tail tip of siphophage T5 : central fibre protein pb4 | | Descriptor: | Probable central straight fiber | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-04-15 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
5JE0
 
 | | Crystal structure of Burkholderia glumae ToxA with bound S-adenosylhomocysteine (SAH) and 1,6-didemethyltoxoflavin | | Descriptor: | Methyl transferase, S-ADENOSYL-L-HOMOCYSTEINE, pyrimido[5,4-e][1,2,4]triazine-5,7(6H,8H)-dione | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
2JSX
 
 | |
1LWE
 
 | | CRYSTAL STRUCTURE OF M41L/T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMN) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
5JE1
 
 | | Crystal structure of Burkholderia glumae ToxA with bound S-adenosylhomocysteine (SAH) and toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, Methyl transferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
7NDT
 
 | | UL40:01 TCR in complex with HLA-E with a non-natural amino acid | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
5JDY
 
 | | Crystal structure of Burkholderia glumae ToxA Y7F mutant with bound S-adenosylhomocysteine (SAH) and toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Methyl transferase, ... | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
7NDU
 
 | | Gag:02 TCR in complex with HLA-E featuring a non-natural amino acid | | Descriptor: | Beta-2-microglobulin, Gag6V(276-284 H4C), HLA class I histocompatibility antigen, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
7NDQ
 
 | | Gag:02 TCR in complex with HLA-E. | | Descriptor: | Beta-2-microglobulin, Gag6V, HLA class I histocompatibility antigen, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
