2YGM
 
 | | THE X-RAY CRYSTAL STRUCTURE OF TANDEM CBM51 MODULES OF SP3GH98, THE FAMILY 98 GLYCOSIDE HYDROLASE FROM STREPTOCOCCUS PNEUMONIAE SP3-BS71, IN COMPLEX WITH THE BLOOD GROUP B ANTIGEN | | Descriptor: | BLOOD GROUP A-AND B-CLEAVING ENDO-BETA-GALACTOSIDASE, CALCIUM ION, SODIUM ION, ... | | Authors: | Higgins, M.A, Ficko-Blean, E, Wright, C, Meloncelli, P.J, Lowary, T.L, Boraston, A.B. | | Deposit date: | 2011-04-19 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Overall Architecture and Receptor Binding of Pneumococcal Carbohydrate Antigen Hydrolyzing Enzymes.
J.Mol.Biol., 411, 2011
|
|
4GFN
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic | | Descriptor: | (1R,5S,6s)-3-[5-chloro-6-ethyl-2-(pyrimidin-5-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]-3-azabicyclo[3.1.0]hexan-6-amine, DNA gyrase subunit B, GLYCEROL | | Authors: | Bensen, D.C, Trzoss, M, Tari, L.W. | | Deposit date: | 2012-08-03 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1T8H
 
 | | 1.8 A CRYSTAL STRUCTURE OF AN UNCHARACTERIZED B. STEAROTHERMOPHILUS PROTEIN | | Descriptor: | BETA-MERCAPTOETHANOL, YlmD protein sequence homologue, ZINC ION | | Authors: | Minasov, G, Shuvalova, L, Mondragon, A, Taneja, B, Moy, S.F, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-12 | | Release date: | 2004-05-18 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A CRYSTAL STRUCTURE OF AN UNCHARACTERIZED B. STEAROTHERMOPHILUS PROTEIN
To be Published
|
|
2RIT
 
 | | Unliganded B-specific-1,3-galactosyltransferase (GTB) | | Descriptor: | GLYCEROL, Glycoprotein-fucosylgalactoside alpha-galactosyltransferase | | Authors: | Evans, S.V, Alfaro, J.A. | | Deposit date: | 2007-10-12 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes.
J.Biol.Chem., 283, 2008
|
|
5DD6
 
 | |
2RIZ
 
 | |
2RVC
 
 | | Solution structure of Zalpha domain of goldfish ZBP-containing protein kinase | | Descriptor: | Interferon-inducible and double-stranded-dependent eIF-2kinase | | Authors: | Lee, A, Park, C, Park, J, Kwon, M, Choi, Y, Kim, K, Choi, B, Lee, J. | | Deposit date: | 2015-07-08 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Z-DNA binding domain of PKR-like protein kinase from Carassius auratus and quantitative analyses of the intermediate complex during B-Z transition.
Nucleic Acids Res., 44, 2016
|
|
2UZT
 
 | | PKA structures of AKT, indazole-pyridine inhibitors | | Descriptor: | (2S)-1-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}-3-PHENYLPROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Zhu, G.D, Gandhi, V.B, Gong, J, Thomas, S, Woods, K.W, Song, X, Li, T, Diebold, R.B, Luo, Y, Liu, X, Guan, R, Klinghofer, V, Johnson, E.F, Bouska, J, Olson, A, Marsh, K.C, Stoll, V.S, Mamo, M, Polakowski, J, Campbell, T.J, Penning, T.D, Li, Q, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Syntheses of Potent, Selective, and Orally Bioavailable Indazole-Pyridine Series of Protein Kinase B/Akt Inhibitors with Reduced Hypotension.
J.Med.Chem., 50, 2007
|
|
2UZU
 
 | | PKA structures of indazole-pyridine series of AKT inhibitors | | Descriptor: | (2S)-1-(1H-INDOL-3-YL)-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Zhu, G.D, Gandhi, V.B, Gong, J, Thomas, S, Woods, K.W, Song, X, Li, T, Diebold, R.B, Luo, Y, Liu, X, Guan, R, Klinghofer, V, Johnson, E.F, Bouska, J, Olson, A, Marsh, K.C, Stoll, V.S, Mamo, M, Polakowski, J, Campbell, T.J, Penning, T.D, Li, Q, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Syntheses of Potent, Selective, and Orally Bioavailable Indazole-Pyridine Series of Protein Kinase B/Akt Inhibitors with Reduced Hypotension.
J.Med.Chem., 50, 2007
|
|
2UZV
 
 | | PKA structures of indazole-pyridine series of AKT inhibitors | | Descriptor: | (2S)-1-[3-(CYCLOHEXYLMETHOXY)PHENYL]-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA,, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Zhu, G.D, Gandhi, V.B, Gong, J, Thomas, S, Woods, K.W, Song, X, Li, T, Diebold, R.B, Luo, Y, Liu, X, Guan, R, Klinghofer, V, Johnson, E.F, Bouska, J, Olson, A, Marsh, K.C, Stoll, V.S, Mamo, M, Polakowski, J, Campbell, T.J, Penning, T.D, Li, Q, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Syntheses of Potent, Selective, and Orally Bioavailable Indazole-Pyridine Series of Protein Kinase B/Akt Inhibitors with Reduced Hypotension.
J.Med.Chem., 50, 2007
|
|
2UZW
 
 | | PKA structures of indazole-pyridine series of AKT inhibitors | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.] PYRAZOLE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Zhu, G.D, Gandhi, V.B, Gong, J, Thomas, S, Woods, K.W, Song, X, Li, T, Diebold, R.B, Luo, Y, Liu, X, Guan, R, Klinghofer, V, Johnson, E.F, Bouska, J, Olson, A, Marsh, K.C, Stoll, V.S, Mamo, M, Polakowski, J, Campbell, T.J, Penning, T.D, Li, Q, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Syntheses of Potent, Selective, and Orally Bioavailable Indazole-Pyridine Series of Protein Kinase B/Akt Inhibitors with Reduced Hypotension.
J.Med.Chem., 50, 2007
|
|
1LPH
 
 | | LYS(B28)PRO(B29)-HUMAN INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Ciszak, E, Beals, J.M, Frank, B.H, Baker, J.C, Carter, N.D, Smith, G.D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of C-terminal B-chain residues in insulin assembly: the structure of hexameric LysB28ProB29-human insulin.
Structure, 3, 1995
|
|
4LV9
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, 7-chloro-3-methyl-2H-1,2,4-benzothiadiazine 1,1-dioxide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2ZJF
 
 | |
4M7W
 
 | | Crystal structure of purine nucleoside phosphorylase from Leptotrichia buccalis C-1013-b, NYSGRC Target 029767. | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-12 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Leptotrichia buccalis C-1013-b, NYSGRC Target 029767.
To be Published
|
|
6CCL
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 1-benzyl-1H-imidazo[4,5-b]pyridine | | Descriptor: | 1-benzyl-1H-imidazo[4,5-b]pyridine, DIMETHYL SULFOXIDE, Phosphopantetheine adenylyltransferase, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
6CT2
 
 | | MYST histone acetyltransferase KAT6A/B in complex with WM-1119 | | Descriptor: | 3-fluoro-N'-[(2-fluorophenyl)sulfonyl]-5-(pyridin-2-yl)benzohydrazide, Histone acetyltransferase KAT8, MAGNESIUM ION, ... | | Authors: | Ren, B, Peat, T.S. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth.
Nature, 560, 2018
|
|
5K9O
 
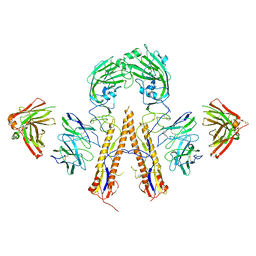 | | Crystal structure of multidonor HV1-18+HD3-9 class broadly neutralizing Influenza A antibody 31.b.09 in complex with Hemagglutinin H1 A/California/04/2009 | | Descriptor: | 31.b.09 Heavy Fv, 31.b.09 Light Fv, Hemagglutinin, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-06-01 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.387 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
3OID
 
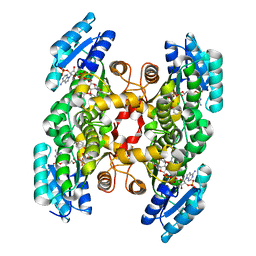 | | Crystal Structure of Enoyl-ACP Reductases III (FabL) from B. subtilis (complex with NADP and TCL) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRICLOSAN | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
6HJD
 
 | | Cholera toxin classical B-pentamer in complex with Lewis-x | | Descriptor: | BICINE, CALCIUM ION, Cholera toxin B subunit, ... | | Authors: | Krengel, U, Heim, J.B. | | Deposit date: | 2018-09-03 | | Release date: | 2019-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structures of cholera toxin in complex with fucosylated receptors point to importance of secondary binding site.
Sci Rep, 9, 2019
|
|
7M6I
 
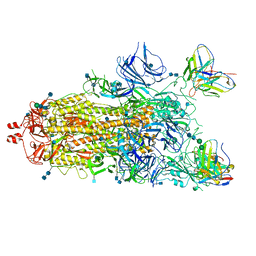 | |
6HMW
 
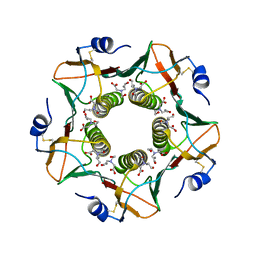 | | Cholera toxin classical B-pentamer in complex with fucose | | Descriptor: | BICINE, CALCIUM ION, Cholera enterotoxin B-subunit, ... | | Authors: | Krengel, U, Heim, J.B. | | Deposit date: | 2018-09-13 | | Release date: | 2019-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of cholera toxin in complex with fucosylated receptors point to importance of secondary binding site.
Sci Rep, 9, 2019
|
|
3M30
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | Deposit date: | 2010-03-08 | | Release date: | 2010-09-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
2O9R
 
 | | beta-glucosidase B complexed with thiocellobiose | | Descriptor: | Beta-glucosidase B, beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose | | Authors: | Isorna, P, Polaina, J, Sanz-Aparicio, J. | | Deposit date: | 2006-12-14 | | Release date: | 2007-10-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
4O2E
 
 | | A peptide complexed with HLA-B*3901 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-39 alpha chain, ... | | Authors: | Sun, M, Liu, J, Qi, J, Tefsen, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | N alpha-terminal acetylation for T cell recognition: molecular basis of MHC class I-restricted n alpha-acetylpeptide presentation
J.Immunol., 192, 2014
|
|
