5Z7B
 
 | |
6GKF
 
 | |
7F2N
 
 | |
6SDF
 
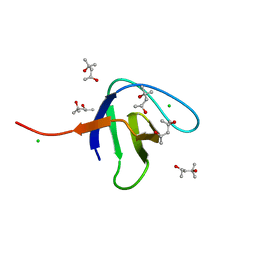 | | N-terminal SH3 domain of Grb2 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Growth factor receptor-bound protein 2 | | Authors: | Bolgov, A.A, Korban, S.A, Luzik, D.A, Rogacheva, O.N, Zhemkov, V.A, Kim, M, Skrynnikov, N.R, Bezprozvanny, I.B. | | Deposit date: | 2019-07-26 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the SH3 domain of growth factor receptor-bound protein 2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6GIM
 
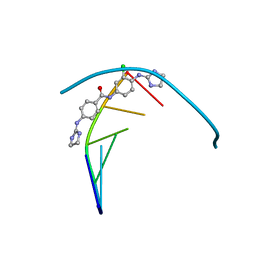 | | Structure of the DNA duplex d(AAATTT)2 with [N-(3-chloro-4-((4,5-dihydro-1H-imidazol-2-yl)amino)phenyl)-4-((4,5-dihydro-1H-imidazol-2- yl)amino)benzamide] - (drug JNI18) | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*T)-3'), MAGNESIUM ION, [4-[(3-chloranyl-4-imidazolidin-2-ylideneazaniumyl-phenyl)carbamoyl]phenyl]-imidazolidin-2-ylidene-azanium | | Authors: | Millan, C.R, Dardonvile, C, de Koning, H.P, Saperas, N, Campos, J.L. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Functional and structural analysis of AT-specific minor groove binders that disrupt DNA-protein interactions and cause disintegration of the Trypanosoma brucei kinetoplast.
Nucleic Acids Res., 45, 2017
|
|
6A8M
 
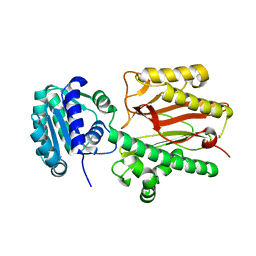 | | N-terminal domain of FACT complex subunit SPT16 from Eremothecium gossypii (Ashbya gossypii) | | Descriptor: | FACT complex subunit SPT16 | | Authors: | Gaur, N.K, Are, V.N, Durani, V, Ghosh, B, Kumar, A, Kulkarni, K, Makde, R.D. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolutionary conservation of protein dynamics: insights from all-atom molecular dynamics simulations of 'peptidase' domain of Spt16.
J.Biomol.Struct.Dyn., 2021
|
|
9DJ8
 
 | | RNA-nsp9 bound to the NiRAN domain of the E-RTC with an empty G-pocket | | Descriptor: | ADENOSINE MONOPHOSPHATE, Non-structural protein 9, RNA-directed RNA polymerase, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2024-09-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | The mechanism for GTP-mediated RNA capping by the SARS-CoV-2 NiRAN domain remains unresolved.
Cell, 188, 2025
|
|
5FUZ
 
 | |
9DIN
 
 | |
5ZUH
 
 | |
9E0O
 
 | | CryoEM structure of inducible Lysine decarboxylase from Hafnia alvei L-hydrazino-Lysine analog at 2.04 Angstrom resolution | | Descriptor: | (2R)-6-amino-2-[(2E)-2-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)hydrazin-1-yl]hexanoic acid, Lysine decarboxylase, inducible | | Authors: | Duhoo, Y, Desfosses, A, Gutsche, I, Doukov, T.I, Berkowitz, D.B. | | Deposit date: | 2024-10-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
5XGT
 
 | |
9E0M
 
 | | CryoEM structure of holoenzyme of inducible Lysine decarboxylase from Hafnia alvei holoenzyme at 2.19 Angstrom resolution | | Descriptor: | Lysine decarboxylase, inducible | | Authors: | Duhoo, Y, Desfosses, A, Gutsche, I, Doukov, T.I, Berkowitz, D.B. | | Deposit date: | 2024-10-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
5SYQ
 
 | | Solution structure of Aquifex aeolicus Aq1974 | | Descriptor: | Uncharacterized protein aq_1974 | | Authors: | Sachleben, J.R, Gawlak, G, Hoey, R.J, Liu, G, Joachimiak, A, Montelione, G.T, Koide, S, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-08-11 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Aromatic claw: A new fold with high aromatic content that evades structural prediction.
Protein Sci., 26, 2017
|
|
5XX5
 
 | | A BPTI-[5,55] variant with C14GA38I mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
9E0Q
 
 | | CryoEM structure of inducible Lysine decarboxylase from Hafnia alvei D-hydrazino-Lysine analog at 2.3 Angstrom resolution | | Descriptor: | (2R)-6-amino-2-[(2E)-2-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)hydrazin-1-yl]hexanoic acid, Lysine decarboxylase, inducible | | Authors: | Duhoo, Y, Desfosses, A, Gutsche, I, Doukov, T.I, Berkowitz, D.B. | | Deposit date: | 2024-10-18 | | Release date: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
1O63
 
 | |
5XX2
 
 | | A BPTI-[5,55] variant with C14GA38L mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
1O6C
 
 | |
5XX4
 
 | | A BPTI-[5,55] variant with C14GA38K mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
6A9U
 
 | | Crystal strcture of Icp55 from Saccharomyces cerevisiae bound to apstatin inhibitor | | Descriptor: | Intermediate cleaving peptidase 55, MANGANESE (II) ION, apstatin | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A9V
 
 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 42 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
9DUI
 
 | |
5XUO
 
 | |
9O38
 
 | | Transmembrane domains of the human sweet receptor (TAS1R2 + TAS1R3) from Class 3 particles (rigidly fitted from PDB:9NOX and 9NOR) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Nanobody 35 (NB35), ... | | Authors: | Juen, Z, Lu, Z, Yu, R, Chang, A.N, Wang, B, Fitzpatrick, A.W.P, Zuker, C.S. | | Deposit date: | 2025-04-06 | | Release date: | 2025-05-14 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The structure of human sweetness.
Cell, 188, 2025
|
|
