3SEU
 
 | | Zn-mediated Polymer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form III) | | Descriptor: | ACETATE ION, GLYCEROL, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SGP
 
 | | Amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-crystallin B chain | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4016 Å) | | Cite: | Atomic view of a toxic amyloid small oligomer.
Science, 335, 2012
|
|
3SGW
 
 | |
3SHL
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V74KL25A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Sue, G, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2011-06-16 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Redirection of internal ionizable residues by engineered cavities
To be Published
|
|
3SKN
 
 | | Crystal structure of the RL42 TCR unliganded | | Descriptor: | RL42 T cell receptor, alpha chain, beta chain | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
3SJV
 
 | | Crystal structure of the RL42 TCR in complex with HLA-B8-FLR | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 3, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
3SEX
 
 | | Ni-mediated Dimer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form II) | | Descriptor: | IODIDE ION, Maltose-binding periplasmic protein, NICKEL (II) ION, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SLJ
 
 | | Pre-cleavage Structure of the Autotransporter EspP - N1023A mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Serine protease espP | | Authors: | Barnard, T.B, Noinaj, N, Easley, N.C, Kuszak, A.J, Buchanan, S.K. | | Deposit date: | 2011-06-24 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter.
J.Mol.Biol., 415, 2012
|
|
3SHT
 
 | |
3SK8
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS M98G apo protein at cryogenic temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, Thermonuclease | | Authors: | Doctrow, B.M, Schlessman, J.L, Garcia-Moreno E, B, Heroux, A. | | Deposit date: | 2011-06-22 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS M98G apo protein at cryogenic temperature
To be Published
|
|
3STN
 
 | | Structure of human LFABP (apo-LFABP) | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Sharma, A, Sharma, A. | | Deposit date: | 2011-07-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Fatty acid induced remodeling within the Human liver fatty acid binding protein
J.Biol.Chem., 2011
|
|
3SP1
 
 | |
3SGR
 
 | | Tandem repeat of amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Tandem repeat of amyloid-related segment of alphaB-crystallin residues 90-100 mutant V91L | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Atomic view of a toxic amyloid small oligomer.
Science, 335, 2012
|
|
3SHQ
 
 | | Crystal Structure of UBLCP1 | | Descriptor: | MAGNESIUM ION, UBLCP1 | | Authors: | Xiao, J, Engel, J.L. | | Deposit date: | 2011-06-16 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SI0
 
 | | Structure of glycosylated human glutaminyl cyclase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D, Stubbs, M.T. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
3SJU
 
 | | Hedamycin Polyketide Ketoreductase bound to NADPH | | Descriptor: | Keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Javidpour, P, Tsai, S.-C. | | Deposit date: | 2011-06-21 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Studies of the Hedamycin Type II Polyketide Ketoreductase (HedKR): Molecular Basis of Stereo- and Regiospecificities.
Biochemistry, 50, 2011
|
|
3SK6
 
 | |
3SKM
 
 | | Crystal structure of the HLA-B8FLRGRAYVL, mutant G8V of the FLR peptide | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 3, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
3SLL
 
 | |
3S5Z
 
 | | Pharmacological Chaperoning in Human alpha-Galactosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, GLYCEROL, ... | | Authors: | Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2011-05-23 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The molecular basis of pharmacological chaperoning in human alpha-galactosidase
Chem.Biol., 18, 2011
|
|
3S6L
 
 | |
3S76
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1H-imidazole-2-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3S73
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3S8Q
 
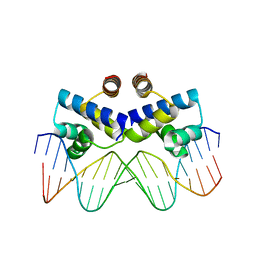 | | Crystal structure of the R-M controller protein C.Esp1396I OL operator complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*AP*CP*TP*TP*AP*TP*AP*GP*TP*CP*CP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*GP*GP*AP*CP*TP*AP*TP*AP*AP*GP*TP*CP*AP*CP*A)-3'), R-M CONTROLLER PROTEIN | | Authors: | McGeehan, J.E, Ball, N.J, Streeter, S.D, Thresh, S.-J, Kneale, G.G. | | Deposit date: | 2011-05-30 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition of dual symmetry by the controller protein C.Esp1396I based on the structure of the transcriptional activation complex.
Nucleic Acids Res., 40, 2012
|
|
3SAP
 
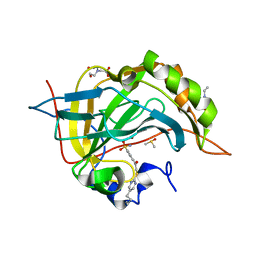 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[(5-butyl-2-pyrimidinyl)sulfanyl]acetyl}benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-{[(5-butylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, BICINE, ... | | Authors: | Grazulis, S, Manakova, E, Tamulaitiene, G. | | Deposit date: | 2011-06-03 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
