7L5H
 
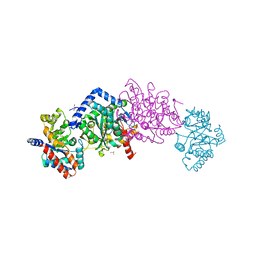 | |
3CD8
 
 | | X-ray Structure of c-Met with triazolopyridazine Inhibitor. | | Descriptor: | 7-methoxy-4-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methoxy]quinoline, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Albrecht, B.K, Harmange, J.-C, Bauer, D, Choquette, D, Dussault, I. | | Deposit date: | 2008-02-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Optimization of Triazolopyridazines as Potent and Selective Inhibitors of the c-Met Kinase.
J.Med.Chem., 51, 2008
|
|
3PMT
 
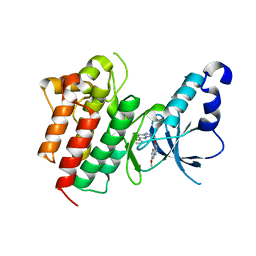
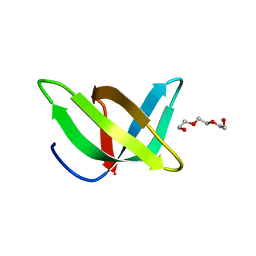 | | Crystal structure of the Tudor domain of human Tudor domain-containing protein 3 | | Descriptor: | TETRAETHYLENE GLYCOL, Tudor domain-containing protein 3 | | Authors: | Lam, R, Bian, C.B, Guo, Y.H, Xu, C, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of TDRD3 and Methyl-Arginine Binding Characterization of TDRD3, SMN and SPF30.
Plos One, 7, 2012
|
|
3NEA
 
 | | Crystal Structure of Peptidyl-tRNA hydrolase from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-tRNA hydrolase | | Authors: | Lam, R, McGrath, T.E, Romanov, V, Gothe, S.A, Peddi, S.R, Razumova, E, Lipman, R.S, Branstrom, A.A, Chirgadze, N.Y. | | Deposit date: | 2010-06-08 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Francisella tularensis peptidyl-tRNA hydrolase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3PGG
 
 | | Crystal structure of cryptosporidium parvum u6 snrna-associated sm-like protein lsm5 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm5. SM domain | | Authors: | Dong, A, Gao, M, Zhao, Y, Lew, J, Wasney, G.A, Kozieradzki, I, Vedadi, M, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-01 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Genome-Scale Protein Expression and Structural Biology of Plasmodium Falciparum and Related Apicomplexan Organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
7MW7
 
 | | Crystal structure of P1G mutant of D-dopachrome tautomerase | | Descriptor: | D-dopachrome decarboxylase, SODIUM ION, SULFATE ION | | Authors: | Manjula, R, Murphy, E.L, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-15 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
4TOY
 
 | |
7MRV
 
 | | F100A mutant structure of MIF2 (D-DT) | | Descriptor: | D-dopachrome decarboxylase, SULFATE ION | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-09 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
2RDG
 
 | | Crystal structure of Staphylococcal Superantigen-Like protein 11 in complex with Sialyl Lewis X | | Descriptor: | CITRIC ACID, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose, POTASSIUM ION, ... | | Authors: | Chung, M.C, Wines, B.D, Baker, H, Langley, R.J, Baker, E.N, Fraser, J.D. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of staphylococcal superantigen-like protein 11 in complex with sialyl Lewis X reveals the mechanism for cell binding and immune inhibition
Mol.Microbiol., 66, 2007
|
|
8GQG
 
 | |
5W56
 
 | | Structure of Apo AztC | | Descriptor: | GLYCEROL, Periplasmic solute binding protein, SODIUM ION | | Authors: | Avalos, D, Yukl, E.T. | | Deposit date: | 2017-06-14 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanisms of zinc binding to the solute-binding protein AztC and transfer from the metallochaperone AztD.
J. Biol. Chem., 292, 2017
|
|
2R5N
 
 | | Crystal structure of transketolase from Escherichia coli in noncovalent complex with acceptor aldose ribose 5-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 5-O-phosphono-beta-D-ribofuranose, CALCIUM ION, ... | | Authors: | Parthier, C, Asztalos, P, Wille, G, Tittmann, K. | | Deposit date: | 2007-09-04 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Strain and Near Attack Conformers in Enzymic Thiamin Catalysis: X-ray Crystallographic Snapshots of Bacterial Transketolase in Covalent Complex with Donor Ketoses Xylulose 5-phosphate and Fructose 6-phosphate, and in Noncovalent Complex with Acceptor Aldose Ribose 5-phosphate.
Biochemistry, 46, 2007
|
|
4OB8
 
 | | Crystal structure of a novel thermostable esterase from Pseudomonas putida ECU1011 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
5E2V
 
 | |
5WBB
 
 | | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins - S112A | | Descriptor: | COPPER (II) ION, GLYCEROL, SULFATE ION, ... | | Authors: | Mann, S.I, Heinisch, T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
5WBL
 
 | |
4OB7
 
 | | Crystal structure of esterase rPPE mutant W187H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
5W90
 
 | | FEZ-1 metallo-beta-lactamase from Legionella gormanii modelled with unknown ligand | | Descriptor: | FEZ-1 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Papamicael, C, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2017-06-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril.
J. Mol. Biol., 325, 2003
|
|
5W97
 
 | | Crystal Structure of CO-bound Cytochrome c Oxidase determined by Serial Femtosecond X-Ray Crystallography at Room Temperature | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Rousseau, D.L, Yeh, S.-R, Ishigami, I, Zatsepin, N.A, Grant, T.D, Fromme, P, Fromme, R. | | Deposit date: | 2017-06-22 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WAB
 
 | |
5WBK
 
 | |
5WJ9
 
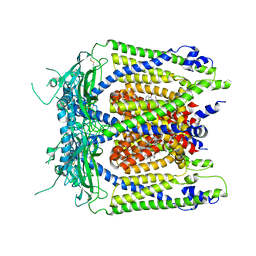 | | Human TRPML1 channel structure in agonist-bound open conformation | | Descriptor: | 2-{2-oxo-2-[(4S)-2,2,4-trimethyl-3,4-dihydroquinolin-1(2H)-yl]ethyl}-1H-isoindole-1,3(2H)-dione, Mucolipin-1 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Human TRPML1 channel structures in open and closed conformations.
Nature, 550, 2017
|
|
5WLL
 
 | |
5WNM
 
 | |
5WKD
 
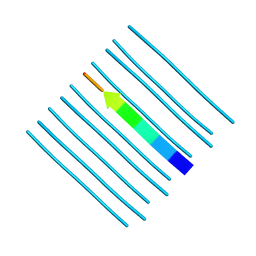 | | Crystal structure of the segment, GNNQGSN, from the low complexity domain of TDP-43, residues 300-306 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Trinh, H, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
