3QGE
 
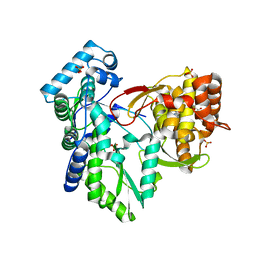 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-N-(4-methoxybenzyl)-1-{[4-(trifluoromethoxy)phenyl]sulfonyl}piperazine-2-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2ARS
 
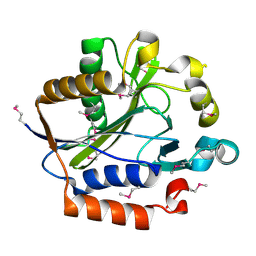 | | Crystal structure of lipoate-protein ligase A From Thermoplasma acidophilum | | Descriptor: | Lipoate-protein ligase A, MAGNESIUM ION | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
2ART
 
 | | Crystal structure of lipoate-protein ligase A bound with lipoyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, LIPOIC ACID, Lipoate-protein ligase A, ... | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
9CHT
 
 | | Human E3 ligase E6AP in complex with HPV16-E6 and p53 | | Descriptor: | Immunoglobulin G-binding protein G/Cellular tumor antigen p53 fusion protein, Protein E6, Ubiquitin-protein ligase E3A | | Authors: | Kenny, S, Das, C. | | Deposit date: | 2024-07-02 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structure of E6AP in complex with HPV16-E6 and p53 reveals a novel ordered domain important for E3 ligase activation.
Structure, 33, 2025
|
|
3A9U
 
 | |
3I5R
 
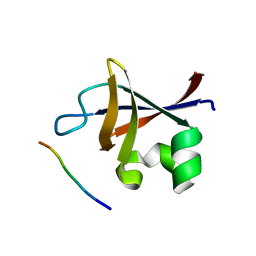 | | PI3K SH3 domain in complex with a peptide ligand | | Descriptor: | Peptide ligand, Phosphatidylinositol 3-kinase regulatory subunit alpha | | Authors: | Batra-Safferling, R, Granzin, J, Modder, S, Hoffmann, S, Willbold, D. | | Deposit date: | 2009-07-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of the phosphatidylinositol 3-kinase (PI3K) SH3 domain in complex with a peptide ligand: role of the anchor residue in ligand binding.
Biol.Chem., 391, 2010
|
|
8QFD
 
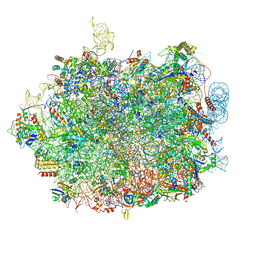 | | UFL1 E3 ligase bound 60S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Makhlouf, L, Kulathu, Y, Zeqiraj, E. | | Deposit date: | 2023-09-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
1X01
 
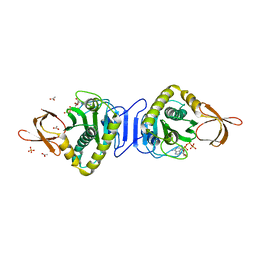 | |
2C7I
 
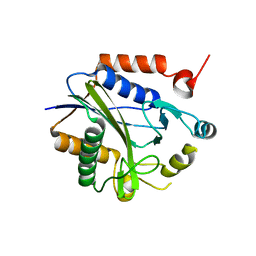 | |
9DRK
 
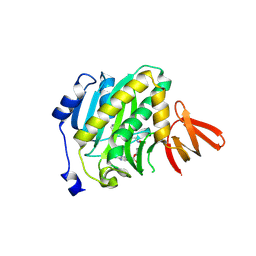 | | Crystal structure of Mycobacterium tuberculosis biotin protein ligase in complex with Bio-1 | | Descriptor: | 5'-deoxy-5'-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}sulfamamido)adenosine, Biotin--[acetyl-CoA-carboxylase] ligase | | Authors: | McCue, W.M, Jayasinghe, Y.P, Aldrich, C.C, Ronning, D.R. | | Deposit date: | 2024-09-25 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Metabolically Stable Adenylation Inhibitors of Biotin Protein Ligase as Antibacterial Agents.
J.Med.Chem., 68, 2025
|
|
9DRN
 
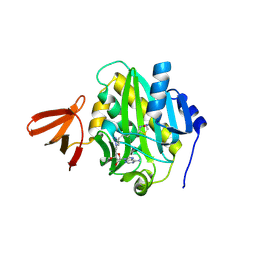 | | Crystal structure of Mycobacterium tuberculosis biotin protein ligase in complex with Bio-4 | | Descriptor: | 5'-deoxy-5'-(4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)adenosine, Biotin--[acetyl-CoA-carboxylase] ligase | | Authors: | McCue, W.M, Jayasinghe, Y.P, Aldrich, C.C, Ronning, D.R. | | Deposit date: | 2024-09-25 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Metabolically Stable Adenylation Inhibitors of Biotin Protein Ligase as Antibacterial Agents.
J.Med.Chem., 68, 2025
|
|
7OYL
 
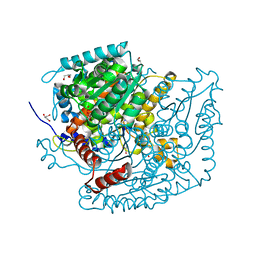 | | Phosphoglucose isomerase of Aspergillus fumigatus in complexed with Glucose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raimi, O.G, Yan, K, Fang, W, van Aalten, D.M.F. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Phosphoglucose Isomerase Is Important for Aspergillus fumigatus Cell Wall Biogenesis.
Mbio, 13, 2022
|
|
2Y66
 
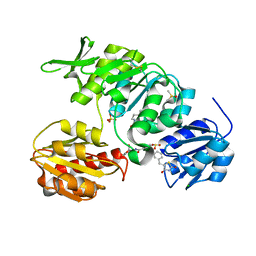 | | New 5-Benzylidenethiazolidine-4-one Inhibitors of Bacterial MurD Ligase: Design, Synthesis, Crystal Structures, and Biological Evaluation | | Descriptor: | (2R)-2-[[3-[[3-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]phenoxy]methyl]phenyl]carbonylamino]pentanedioic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Zidar, N, Tomasic, T, Sink, R, Kovac, A, Patin, D, Blanot, D, Contreras-Martel, C, Dessen, A, Muller-Premru, M, Zega, A, Gobec, S, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2011-01-20 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | New 5-Benzylidenethiazolidin-4-One Inhibitors of Bacterial Murd Ligase: Design, Synthesis, Crystal Structures, and Biological Evaluation.
Eur.J.Med.Chem, 46, 2011
|
|
6OJB
 
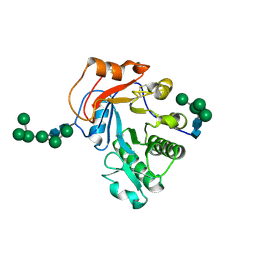 | | Crystal Structure of Aspergillus fumigatus Ega3 complex with galactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-alpha-D-galactopyranose, ... | | Authors: | Bamford, N.C, Howell, P.L. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
1WAT
 
 | |
1WAS
 
 | |
6OJ1
 
 | | Crystal Structure of Aspergillus fumigatus Ega3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bamford, N.C, Subramanian, A.S, Millan, C, Uson, I, Howell, P.L. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
2Y67
 
 | | New 5-Benzylidenethiazolidine-4-one Inhibitors of Bacterial MurD Ligase: Design, Synthesis, Crystal Structures, and Biological Evaluation | | Descriptor: | (2R)-2-[[4-[[4-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]phenoxy]methyl]phenyl]sulfonylamino]pentanedioic acid, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Zidar, N, Tomasic, T, Sink, R, Kovac, A, Patin, D, Blanot, D, Contreras-Martel, C, Dessen, A, Muller-Premru, M, Zega, A, Gobec, S, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2011-01-20 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New 5-Benzylidenethiazolidin-4-One Inhibitors of Bacterial Murd Ligase: Design, Synthesis, Crystal Structures, and Biological Evaluation.
Eur.J.Med.Chem, 46, 2011
|
|
1XDN
 
 | | High resolution crystal structure of an editosome enzyme from trypanosoma brucei: RNA editing ligase 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA editing ligase MP52 | | Authors: | Deng, J, Schnaufer, A, Salavati, R, Stuart, K.D, Hol, W.G. | | Deposit date: | 2004-09-07 | | Release date: | 2004-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1.
J.Mol.Biol., 343, 2004
|
|
4HTO
 
 | |
2DXU
 
 | | Crystal Structure Of Biotin Protein Ligase From Pyrococcus Horikoshii Complexed with Biotinyl-5'-AMP, Mutation R48A | | Descriptor: | BIOTINYL-5-AMP, biotin--[acetyl-CoA-carboxylase] ligase | | Authors: | Bagautdinov, B, Taketa, M, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-30 | | Release date: | 2007-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate
J.Biol.Chem., 283, 2008
|
|
2FN9
 
 | | Thermotoga maritima Ribose Binding Protein Unliganded Form | | Descriptor: | ribose ABC transporter, periplasmic ribose-binding protein | | Authors: | Cuneo, M.J, Changela, A, Tian, Y, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2006-01-10 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ligand-induced conformational changes in a thermophilic ribose-binding protein.
Bmc Struct.Biol., 8, 2008
|
|
1OQE
 
 | | Crystal structure of sTALL-1 with BAFF-R | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
2H0D
 
 | |
8BI4
 
 | |
