1C9Z
 
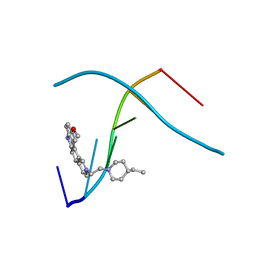 | | D232-CGTACG | | Descriptor: | 1,3-DI[[[10-METHOXY-7H-PYRIDO[4,3-C]CARBAZOL-2-IUMYL]-ETHYL]-PIPERIDIN-4-YL]-PROPANE, 5'-D(*CP*GP*TP*AP*CP*G)-3' | | Authors: | Williams, L.D. | | Deposit date: | 1999-08-03 | | Release date: | 2000-04-02 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effects of cationic charge on three-dimensional structures of intercalative complexes: structure of a bis-intercalated DNA complex solved by MAD phasing.
Curr.Med.Chem., 7, 2000
|
|
1CX1
 
 | | SECOND N-TERMINAL CELLULOSE-BINDING DOMAIN FROM CELLULOMONAS FIMI BETA-1,4-GLUCANASE C, NMR, 22 STRUCTURES | | Descriptor: | ENDOGLUCANASE C | | Authors: | Brun, E, Johnson, P.E, Creagh, L.A, Haynes, C.A, Tomme, P, Webster, P, Kilburn, D.G, McIntosh, L.P. | | Deposit date: | 1999-08-27 | | Release date: | 2000-04-02 | | Last modified: | 2017-02-01 | | Method: | SOLUTION NMR | | Cite: | Structure and binding specificity of the second N-terminal cellulose-binding domain from Cellulomonas fimi endoglucanase C.
Biochemistry, 39, 2000
|
|
1QAJ
 
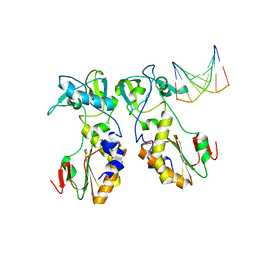 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3'), REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-03-18 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain.
J.Mol.Biol., 296, 2000
|
|
1D5S
 
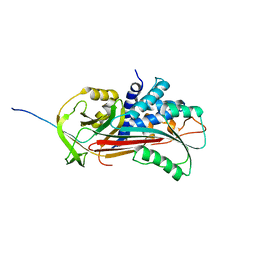 | | CRYSTAL STRUCTURE OF CLEAVED ANTITRYPSIN POLYMER | | Descriptor: | P1-ARG ANTITRYPSIN | | Authors: | Dunstone, M.A, Dai, W, Whisstock, J.C, Rossjohn, J, Pike, R.N, Feil, S.C, Le Bonneic, B.F, Parker, M.W, Bottomley, S.P. | | Deposit date: | 1999-10-11 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cleaved antitrypsin polymers at atomic resolution.
Protein Sci., 9, 2000
|
|
1DTT
 
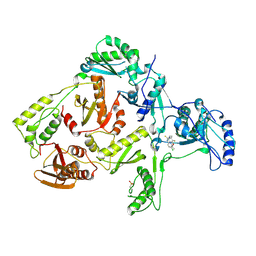 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 (PETT130A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-04-02 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1EEG
 
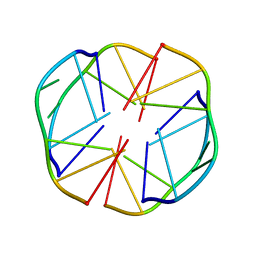 | | A(GGGG)A HEXAD PAIRING ALIGMENT FOR THE D(G-G-A-G-G-A-G) SEQUENCE | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*A)-3') | | Authors: | Kettani, A, Gorin, A, Majumdar, A, Hermann, T, Skripkin, E, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations.
J.Mol.Biol., 297, 2000
|
|
1EOO
 
 | | ECORV BOUND TO COGNATE DNA | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*AP*TP*AP*TP*CP*TP*TP*C)-3'), TYPE II RESTRICTION ENZYME ECORV | | Authors: | Horton, N.C, Perona, J.J. | | Deposit date: | 2000-03-23 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystallographic snapshots along a protein-induced DNA-bending pathway.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EN1
 
 | | STRUCTURE OF THE HIV-1 MINUS STRAND PRIMER BINDING SITE | | Descriptor: | DNA (5'-D(P*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3') | | Authors: | Johnson, P.E, Turner, R.B, Wu, Z.R, Levin, J.G, Summers, M.F. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription
Biochemistry, 39, 2000
|
|
1EOP
 
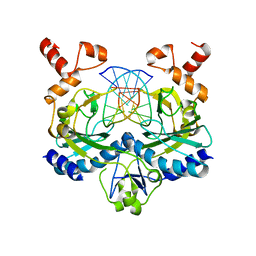 | | ECORV BOUND TO COGNATE DNA | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*AP*TP*AP*TP*CP*TP*TP*C)-3'), TYPE II RESTRICTION ENZYME ECORV | | Authors: | Horton, N.C, Perona, J.J. | | Deposit date: | 2000-03-23 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic snapshots along a protein-induced DNA-bending pathway.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EMQ
 
 | |
1D0E
 
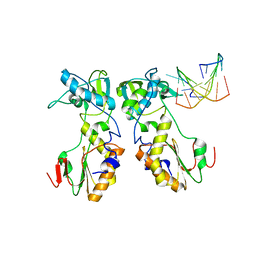 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | 5'-D(*TP*TP*TP*CP*AP*TP*GP*CP*AP*TP*G)-3', REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M.L, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-09-09 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain
J.Mol.Biol., 296, 2000
|
|
1DKR
 
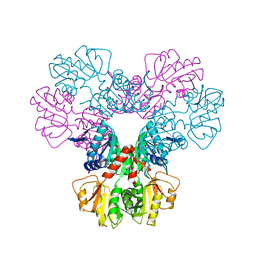 | | CRYSTAL STRUCTURES OF BACILLUS SUBTILIS PHOSPHORIBOSYLPYROPHOSPHATE SYNTHETASE: MOLECULAR BASIS OF ALLOSTERIC INHIBITION AND ACTIVATION. | | Descriptor: | PHOSPHORIBOSYL PYROPHOSPHATE SYNTHETASE, SULFATE ION | | Authors: | Eriksen, T.A, Kadziola, A, Bentsen, A.-K, Harlow, K.W, Larsen, S. | | Deposit date: | 1999-12-08 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase.
Nat.Struct.Biol., 7, 2000
|
|
1DKU
 
 | | CRYSTAL STRUCTURES OF BACILLUS SUBTILIS PHOSPHORIBOSYLPYROPHOSPHATE SYNTHETASE: MOLECULAR BASIS OF ALLOSTERIC INHIBITION AND ACTIVATION. | | Descriptor: | METHYL PHOSPHONIC ACID ADENOSINE ESTER, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PROTEIN (PHOSPHORIBOSYL PYROPHOSPHATE SYNTHETASE) | | Authors: | Eriksen, T.A, Kadziola, A, Bentsen, A.-K, Harlow, K.W, Larsen, S. | | Deposit date: | 1999-12-08 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase.
Nat.Struct.Biol., 7, 2000
|
|
1DJM
 
 | | SOLUTION STRUCTURE OF BEF3-ACTIVATED CHEY FROM ESCHERICHIA COLI | | Descriptor: | CHEMOTAXIS PROTEIN Y | | Authors: | Cho, H.S, Lee, S.Y, Yan, D, Pan, X, Parkinson, J.S, Kustu, S, Wemmer, D.E, Pelton, J.G. | | Deposit date: | 1999-12-03 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of activated CheY.
J.Mol.Biol., 297, 2000
|
|
1EHD
 
 | |
1EFD
 
 | | PERIPLASMIC FERRIC SIDEROPHORE BINDING PROTEIN FHUD COMPLEXED WITH GALLICHROME | | Descriptor: | FERRICHROME-BINDING PERIPLASMIC PROTEIN, GALLICHROME | | Authors: | Clarke, T.E, Ku, S.-Y, Dougan, D.R, Vogel, H.J, Tari, L.W. | | Deposit date: | 2000-02-07 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the ferric siderophore binding protein FhuD complexed with gallichrome.
Nat.Struct.Biol., 7, 2000
|
|
1EHH
 
 | |
1EJG
 
 | | CRAMBIN AT ULTRA-HIGH RESOLUTION: VALENCE ELECTRON DENSITY. | | Descriptor: | CRAMBIN (PRO22,SER22/LEU25,ILE25) | | Authors: | Jelsch, C, Teeter, M.M, Lamzin, V, Pichon-Lesme, V, Blessing, B, Lecomte, C. | | Deposit date: | 2000-03-02 | | Release date: | 2000-04-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (0.54 Å) | | Cite: | Accurate protein crystallography at ultra-high resolution: valence electron distribution in crambin.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1C2P
 
 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Lesburg, C.A, Cable, M.B, Ferrari, E, Hong, Z, Mannarino, A.F, Weber, P.C. | | Deposit date: | 1999-07-26 | | Release date: | 2000-04-05 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site.
Nat.Struct.Biol., 6, 1999
|
|
1BV8
 
 | |
1DVJ
 
 | | CRYSTAL STRUCTURE OF OROTIDINE MONOPHOSPHATE DECARBOXYLASE COMPLEXED WITH 6-AZAUMP | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, OROTIDINE 5'-PHOSPHATE DECARBOXYLASE | | Authors: | Wu, N, Mo, Y, Gao, J, Pai, E.F. | | Deposit date: | 2000-03-30 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Electrostatic stress in catalysis: structure and mechanism of the enzyme orotidine monophosphate decarboxylase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1CIV
 
 | | CHLOROPLAST NADP-DEPENDENT MALATE DEHYDROGENASE FROM FLAVERIA BIDENTIS | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-MALATE DEHYDROGENASE | | Authors: | Carr, P.D, Ashton, A.R, Verger, D, Ollis, D.L. | | Deposit date: | 1999-04-01 | | Release date: | 2000-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chloroplast NADP-malate dehydrogenase: structural basis of light-dependent regulation of activity by thiol oxidation and reduction.
Structure Fold.Des., 7, 1999
|
|
1D2R
 
 | |
1DV7
 
 | |
1DVK
 
 | |
