2JRO
 
 | | Solution NMR Structure of SO0334 from Shewanella oneidensis. Northeast Structural Genomics Target SoR75 | | Descriptor: | Uncharacterized protein | | Authors: | Tang, Y, Wang, D, Nwosu, C, Cunningham, K, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-28 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of SO0334 from Shewanella oneidensis.
To be Published
|
|
1WTS
 
 | |
2JZW
 
 | | How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription | | Descriptor: | DNA (5'-D(*DGP*DTP*DCP*DCP*DCP*DTP*DGP*DTP*DTP*DCP*DGP*DGP*DGP*DC)-3'), HIV-1 nucleocapsid protein NCp7(12-55), ZINC ION | | Authors: | Bourbigot, S, Ramalanjaona, N, Salgado, G.F.J, Mely, Y, Roques, B.P, Bouaziz, S, Morellet, N. | | Deposit date: | 2008-01-21 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription.
J.Mol.Biol., 383, 2008
|
|
2K3C
 
 | | Structural and Functional Characterization of TM IX of the NHE1 Isoform of the Na+/H+ Exchanger | | Descriptor: | TMIX peptide | | Authors: | Reddy, T, Ding, J, Li, X, Sykes, B.D, Fliegel, L, Rainey, J.K. | | Deposit date: | 2008-05-01 | | Release date: | 2008-06-03 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Transmembrane Segment IX of the NHE1 Isoform of the Na+/H+ Exchanger.
J.Biol.Chem., 283, 2008
|
|
2KLW
 
 | |
2KT6
 
 | | Structural homology between the C-terminal domain of the PapC usher and its plug | | Descriptor: | Outer membrane usher protein papC | | Authors: | Ford, B, Rego, A, Ragan, T.J, Pinkner, J, Dodson, K, Driscoll, P.C, Hultgren, S, Waksman, G. | | Deposit date: | 2010-01-19 | | Release date: | 2010-04-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural Homology between the C-Terminal Domain of the PapC Usher and Its Plug.
J.Bacteriol., 192, 2010
|
|
2KE0
 
 | | Solution structure of peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Zheng, S, Leeper, T, Napuli, A, Nakazawa, S.H, Varani, G, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structure of a Burkholderia pseudomallei immunophilin-inhibitor complex reveals new approaches to antimicrobial development.
Biochem.J., 437, 2011
|
|
1XBD
 
 | | INTERNAL XYLAN BINDING DOMAIN FROM CELLULOMONAS FIMI XYLANASE D, NMR, 5 STRUCTURES | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Bolam, D.N, Cooper, A, Ciruela, A, Hazlewood, G.P, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 1998-10-16 | | Release date: | 1999-07-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity.
Structure Fold.Des., 7, 1999
|
|
2H3A
 
 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2K5U
 
 | |
207D
 
 | | SOLUTION STRUCTURE OF MITHRAMYCIN DIMERS BOUND TO PARTIALLY OVERLAPPING SITES ON DNA | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYROXY-7-METHYLANTHRACENE, 2,6-dideoxy-3-C-methyl-beta-D-ribo-hexopyranose-(1-3)-2,6-dideoxy-beta-D-galactopyranose-(1-3)-beta-D-Olivopyranose, DNA (5'-D(*TP*AP*GP*CP*TP*AP*GP*CP*TP*A)-3'), ... | | Authors: | Sastry, M, Fiala, R, Patel, D.J. | | Deposit date: | 1995-04-20 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mithramycin dimers bound to partially overlapping sites on DNA.
J.Mol.Biol., 251, 1995
|
|
2KSQ
 
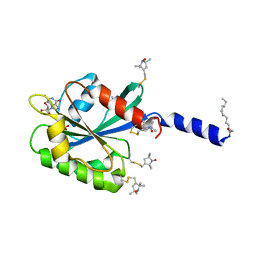 | | The myristoylated yeast ARF1 in a GTP and bicelle bound conformation | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Liu, Y, Kahn, R, Prestegard, J. | | Deposit date: | 2010-01-12 | | Release date: | 2010-07-07 | | Last modified: | 2011-07-27 | | Method: | SOLUTION NMR | | Cite: | Dynamic structure of membrane-anchored Arf*GTP.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2H3C
 
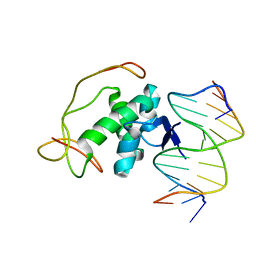 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2KQY
 
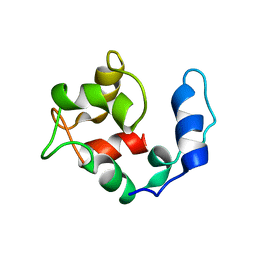 | | Solution structure of Avian Thymic Hormone | | Descriptor: | Parvalbumin, thymic | | Authors: | Henzl, M.T. | | Deposit date: | 2009-11-24 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of Avian Thymic Hormone, a High-Affinity Avian beta-Parvalbumin, in the Ca(2+)-Free and Ca(2+)-Bound States.
J.Mol.Biol., 397, 2010
|
|
2KO7
 
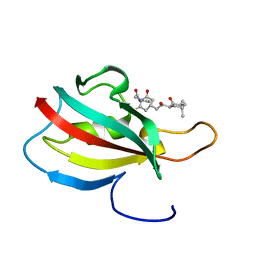 | | Solution structure of peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei complexed with Cycloheximide-N-ethylethanoate | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, ethyl (4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}-2,6-dioxopiperidin-1-yl)acetate | | Authors: | Zheng, S, Leeper, T, Varani, G, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-09-11 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structure of a Burkholderia pseudomallei immunophilin-inhibitor complex reveals new approaches to antimicrobial development.
Biochem.J., 437, 2011
|
|
2KXL
 
 | |
2KSN
 
 | | Solution Structure of the N-terminal Domain of DC-UbP/UBTD2 | | Descriptor: | Ubiquitin domain-containing protein 2 | | Authors: | Song, A, Zhou, C, Guan, X, Sze, K, Hu, H. | | Deposit date: | 2010-01-08 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of DC-UbP/UBTD2 and its interaction with ubiquitin
Protein Sci., 19, 2010
|
|
2KXA
 
 | |
2KT2
 
 | | Structure of NmerA, the N-terminal HMA domain of Tn501 Mercuric Reductase | | Descriptor: | Mercuric reductase | | Authors: | Ledwidge, R, Danacea, F, Dotsch, V, Miller, S.M. | | Deposit date: | 2010-01-17 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NmerA of Tn501 mercuric ion reductase: structural modulation of the pKa values of the metal binding cysteine thiols.
Biochemistry, 49, 2010
|
|
2KT3
 
 | | Structure of Hg-NmerA, Hg(II) complex of the N-terminal domain of Tn501 Mercuric Reductase | | Descriptor: | MERCURY (II) ION, Mercuric reductase | | Authors: | Miller, S.M, Ledwidge, R, Danacea, F, Dotsch, V. | | Deposit date: | 2010-01-17 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NmerA of Tn501 mercuric ion reductase: structural modulation of the pKa values of the metal binding cysteine thiols.
Biochemistry, 49, 2010
|
|
2JNT
 
 | | Structure of Bombyx mori Chemosensory Protein 1 in Solution | | Descriptor: | Chemosensory protein CSP1 | | Authors: | Jansen, S, Zidek, L, Chmelik, J, Novak, P, Padrta, P, Picimbon, J, Lofstedt, C, Sklenar, V. | | Deposit date: | 2007-02-02 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of Bombyx mori chemosensory protein 1 in solution
Arch.Insect Biochem.Physiol., 66, 2007
|
|
2JPJ
 
 | | Lactococcin G-a in DPC | | Descriptor: | Bacteriocin lactococcin-G subunit alpha | | Authors: | Rogne, P, Fimland, G, Nissen-Meyer, J, Kristiansen, P.E. | | Deposit date: | 2007-05-15 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin lactococcin G
Biochim.Biophys.Acta, 1784, 2008
|
|
2JXV
 
 | |
2JYG
 
 | |
2LSF
 
 | | Structure and Stability of Duplex DNA Containing (5'S) 5',8-Cyclo-2'-Deoxyadenosine: An Oxidative Lesion Repair by NER | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*(02I)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Zaliznyak, T, de los Santos, C, Lukin, M. | | Deposit date: | 2012-04-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Stability of Duplex DNA Containing (5'S)-5',8-Cyclo-2'-deoxyadenosine: An Oxidatively Generated Lesion Repaired by NER.
Chem.Res.Toxicol., 25, 2012
|
|
