1TG0
 
 | |
4E0K
 
 | | Crystal Structure of the amyloid-fibril forming peptide KDWSFY derived from human Beta 2 Microglobulin (58-63) | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Amyloidogenic peptide segment KDWSFY, SODIUM ION | | Authors: | Zhao, M, Liu, C, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3DW4
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-OCH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3X0K
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ES-state at 0.97 angstrom resolution | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-16 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
485D
 
 | |
4KXV
 
 | | Human transketolase in covalent complex with donor ketose D-xylulose-5-phosphate, crystal 1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, D-XYLITOL-5-PHOSPHATE, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
8B34
 
 | |
6RII
 
 | | Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by fluoride anions at sub-atomic resolution. Fourth structure of the series with 1200 KGy dose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, FLUORIDE ION, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2019-04-24 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: insights into the enzymatic reaction mechanism.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7TH6
 
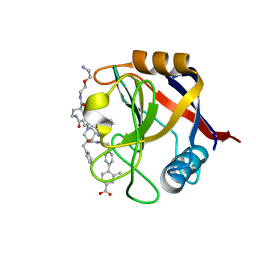 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B21 | | Descriptor: | 4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}-2-methyl[1,1'-biphenyl]-4-carboxylic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
4ACJ
 
 | | Crystal structure of the TLDC domain of Oxidation resistance protein 2 from zebrafish | | Descriptor: | WU:FB25H12 PROTEIN, | | Authors: | Blaise, M, B Alsarraf, H.M.A, Wong, J.E.M.M, Midtgaard, S.R, Laroche, F, Schack, L, Spaink, H, Stougaard, J, Thirup, S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal Structure of the Tldc Domain of Oxidation Resistance Protein 2 from Zebrafish.
Proteins, 80, 2012
|
|
5JEU
 
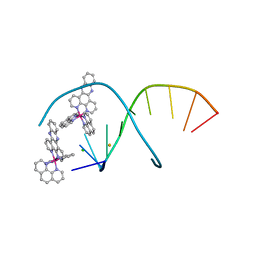 | | del-[Ru(phen)2(dppz)]2+ bound to d(TCGGCGCCGA) with Ba2+ | | Descriptor: | BARIUM ION, CHLORIDE ION, DNA (5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3'), ... | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2016-04-19 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Delta chirality ruthenium 'light-switch' complexes can bind in the minor groove of DNA with five different binding modes.
Nucleic Acids Res., 44, 2016
|
|
2BF6
 
 | |
5RE1
 
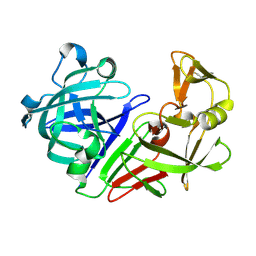 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 58 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
7FYV
 
 | | Crystal Structure of human FABP4 in complex with 5-(4-chlorophenyl)-9-methyl-3-oxa-4-azatricyclo[5.2.1.02,6]dec-4-ene-8-carboxylic acid | | Descriptor: | (3aR,4R,5R,6S,7R,7aR)-3-(4-chlorophenyl)-6-methyl-3a,4,5,6,7,7a-hexahydro-4,7-methano-1,2-benzoxazole-5-carboxylic acid, 1,2-ETHANEDIOL, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Wickens, J, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
6RHH
 
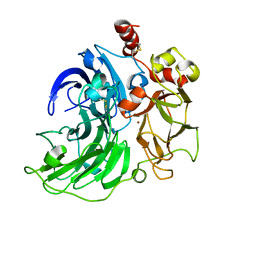 | | Single crystal serial study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi at sub-atomic resolution. Third structure of the series with 315 KGy dose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Laccase 2, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: insights into the enzymatic reaction mechanism.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4PNO
 
 | |
3LL2
 
 | |
6TD8
 
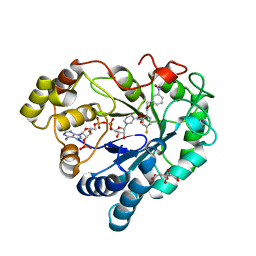 | | Human Aldose Reductase Mutant L301A in Complex with a Ligand with an IDD Structure ({5-fluoro-2-[(3-nitrobenzyl)carbamoyl]phenoxy}acetic acid) | | Descriptor: | Aldo-keto reductase family 1 member B1, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubert, L.-S, Ley, M, Heine, A, Klebe, G. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Human Aldose Reductase Mutant L301A in Complex with a Ligand with an IDD Structure ({5-fluoro-2-[(3-nitrobenzyl)carbamoyl]phenoxy}acetic acid)
To Be Published
|
|
6HMQ
 
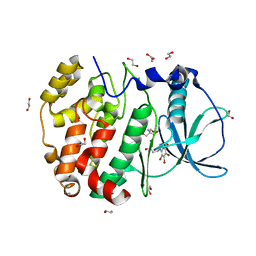 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE BENZOTRIAZOLE-TYPE INHIBITOR MB002 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(4,5,6,7-tetrabromo-1H-benzotriazol-1-yl)propan-1-ol, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
8B97
 
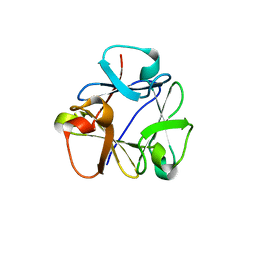 | | N-terminal beta-trefoil lectin domain of the Laccaria bicolor lectin in complex with N-acetyl-lactosamine | | Descriptor: | Beta-trefoil domain of the LBL lectin, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Acebron, I, Campanero-Rhodes, M.A, Solis, D, Menendez, M, Garcia, C, Lillo, M.P, Mancheno, J.M. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Atomic crystal structure and sugar specificity of a beta-trefoil lectin domain from the ectomycorrhizal basidiomycete Laccaria bicolor.
Int.J.Biol.Macromol., 233, 2023
|
|
5RCS
 
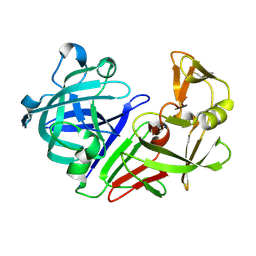 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 14 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
7L71
 
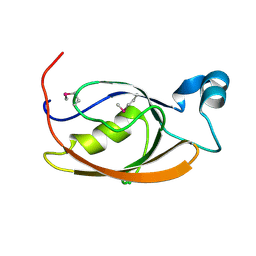 | |
5RDF
 
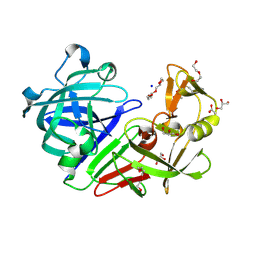 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 37 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
4MTU
 
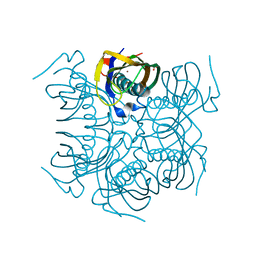 | | beta-Alanyl-CoA:Ammonia Lyase from Clostridium propionicum | | Descriptor: | Beta-alanyl-CoA:ammonia lyase 2, SULFATE ION, ZINC ION | | Authors: | Heine, A, Reuter, K. | | Deposit date: | 2013-09-20 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | High resolution crystal structure of Clostridium propionicum beta-alanyl-CoA:ammonia lyase, a new member of the "hot dog fold" protein superfamily.
Proteins, 82, 2014
|
|
5RDP
 
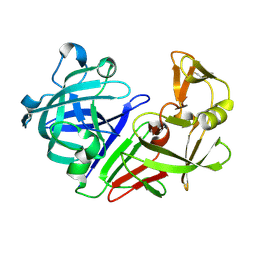 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 41 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
