1S1K
 
 | |
3IXN
 
 | |
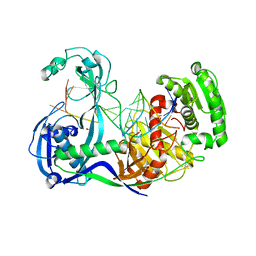
3F73
 
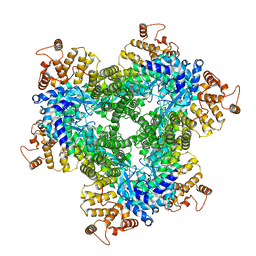
 | | Alignment of guide-target seed duplex within an argonaute silencing complex | | Descriptor: | ARGONAUTE, DNA (5'-D(P*DTP*DGP*DAP*DGP*DGP*DTP*DAP*DGP*DTP*DAP*DGP*DGP*DTP*DTP*DGP*DTP*DA*DTP*DAP*DGP*DT)-3'), MAGNESIUM ION, ... | | Authors: | Wang, Y, Li, H, Sheng, G, Juranek, S, Tuschl, T, Patel, D.J. | | Deposit date: | 2008-11-07 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex.
Nature, 456, 2008
|
|
7Z0Z
 
 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis, Y44F variant | | Descriptor: | AbiK | | Authors: | Figiel, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W, Nowotny, M. | | Deposit date: | 2022-02-24 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
1EKI
 
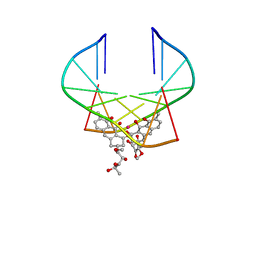 | | AVERAGE SOLUTION STRUCTURE OF D(TTGGCCAA)2 BOUND TO CHROMOMYCIN-A3 AND COBALT | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-4-O-acetyl-2,6-dideoxy-beta-D-galactopyranose, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-beta-D-Olivopyranose-(1-3)-beta-D-Olivopyranose, ... | | Authors: | Gochin, M. | | Deposit date: | 2000-03-08 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance.
Structure Fold.Des., 8, 2000
|
|
6V5K
 
 | |
6PZ3
 
 | |
3HWU
 
 | |
1N17
 
 | | Structure and Dynamics of Thioguanine-modified Duplex DNA | | Descriptor: | 5'-D(*GP*CP*TP*AP*AP*GP*(S6G)P*AP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*TP*CP*CP*TP*TP*AP*GP*C)-3' | | Authors: | Somerville, L, Krynetski, E.Y, Krynetskaia, N.F, Beger, R.D, Zhang, W, Marhefka, C.A, Evans, W.E, Kriwacki, R.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of thioguanine-modified duplex DNA
J.Biol.Chem., 278, 2003
|
|
1N1O
 
 | | Crystal Structure of a B-form DNA Duplex Containing (L)-alpha-threofuranosyl (3'-2') Nucleosides: A Four-Carbon Sugar is Easily Accommodated into the Backbone of DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*(TFT)P*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Wilds, C.J, Wawrzak, Z, Krishnamurthy, R, Eschenmoser, A, Egli, M. | | Deposit date: | 2002-10-18 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of a B-Form DNA Duplex Containing (L)-alpha-Threofuranosyl (3'-->2') Nucleosides: A
Four-Carbon Sugar Is Easily Accommodated into the Backbone of DNA
J.Am.Chem.Soc., 124, 2002
|
|
5V0Q
 
 | |
1R68
 
 | | Role of the amino sugar in DNA binding of disaccharide anthracyclines: crystal structure of MAR70/d(CGATCG) complex | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Temperini, C, Cirilli, M, Aschi, M, Ughetto, G. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Role of the amino sugar in DNA binding of disaccharide anthracyclines: crystal structure of the complex MAR70/d(CGATCG).
BIOORG.MED.CHEM., 13, 2005
|
|
7ART
 
 | | 48 helix bundle DNA origami brick | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Feigl, E, Kube, M, Kohler, F, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
2W58
 
 | | Crystal Structure of the DnaI | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, K.L, Lo, Y.H, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Interplay between the Replicative Helicase Dnac and its Loader Protein Dnai from Geobacillus Kaustophilus.
J.Mol.Biol., 393, 2009
|
|
1RVE
 
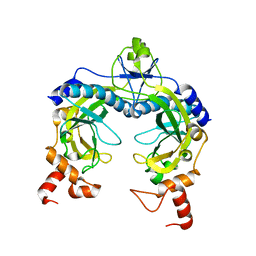 | |
2IQC
 
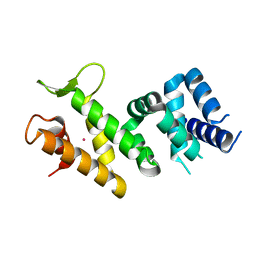 | | Crystal structure of Human FancF Protein that Functions in the Assembly of a DNA Damage Signaling Complex | | Descriptor: | Fanconi anemia group F protein, MERCURY (II) ION | | Authors: | Kowal, P, Gurtan, A.M, Stuckert, P, Lehmann, C, D'Andrea, A, Ellenberger, T.E. | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex.
J.Biol.Chem., 282, 2007
|
|
3FK2
 
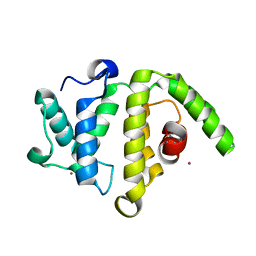 | | Crystal structure of the RhoGAP domain of human glucocorticoid receptor DNA-binding factor 1 | | Descriptor: | Glucocorticoid receptor DNA-binding factor 1, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-15 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the RhoGAP domain of human glucocorticoid receptor DNA-binding factor 1
To be Published
|
|
3EY1
 
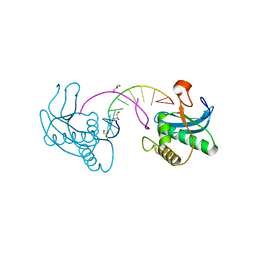 | |
3GO3
 
 | | Interactions of an echinomycin-DNA complex with manganese(II) ions | | Descriptor: | 2-CARBOXYQUINOXALINE, 5'-D(*AP*CP*GP*TP*AP*CP*GP*T)-3', DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pfoh, R, Cuesta-Seijo, J.A, Sheldrick, G.M. | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-31 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Interaction of an Echinomycin-DNA Complex with Manganese Ion
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5CKI
 
 | | Crystal structure of 9DB1* deoxyribozyme (Cobalt hexammine soaked crystals) | | Descriptor: | COBALT (II) ION, DNA (44-MER), MAGNESIUM ION, ... | | Authors: | Ponce-Salvatierra, A, Hoebartner, C, Pena, V. | | Deposit date: | 2015-07-15 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal structure of a DNA catalyst.
Nature, 529, 2016
|
|
1P96
 
 | | Solution Structure of a Wedge-Shaped Synthetic Molecule at a Two-Base Bulge Site in DNA | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*AP*GP*TP*TP*CP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*GP*AP*TP*GP*CP*GP*TP*G)-3', SPIRO[NAPHTHALENE-2(3H),3'(10'H)-PENTALENO[1,2-B]NAPHTHALENE]-3,10'-DIONE, ... | | Authors: | Hwang, G.S, Jones, G.B, Goldberg, I.H. | | Deposit date: | 2003-05-09 | | Release date: | 2003-08-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Wedge-Shaped Synthetic Molecule at a Two-Base Bulge Site in DNA
Biochemistry, 42, 2003
|
|
1DNP
 
 | | STRUCTURE OF DEOXYRIBODIPYRIMIDINE PHOTOLYASE | | Descriptor: | 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Park, H.-W, Sancar, A, Deisenhofer, J. | | Deposit date: | 1995-07-03 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of DNA photolyase from Escherichia coli.
Science, 268, 1995
|
|
8AYG
 
 | | Crystal structure of an intramolecular i-motif at the insulin-linked polymorphic region (ILPR) | | Descriptor: | Insulin-linked polymorphic region, ILPR DNA (31-MER) | | Authors: | Parkinson, G.N, Alexandrou, E, Waller, Z.A.E, El-Omari, K. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into i-motif DNA structures in sequences from the insulin-linked polymorphic region.
Nat Commun, 15, 2024
|
|
3FQB
 
 | |
8G2P
 
 | |
