8J19
 
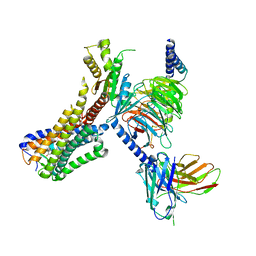 | | Cryo-EM structure of the LY237-bound GPR84 receptor-Gi complex | | Descriptor: | 6-nonylpyridine-2,4-diol, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J18
 
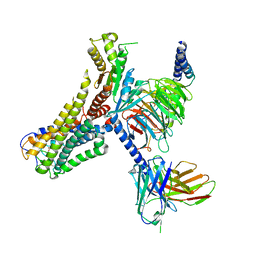 | | Cryo-EM structure of the 3-OH-C12-bound GPR84 receptor-Gi complex | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
6HN0
 
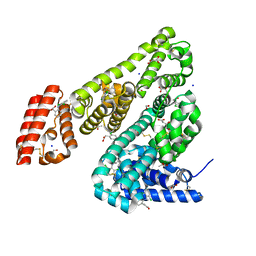 | | Complex of Ovine Serum Albumin with diclofenac | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ACETATE ION, ... | | Authors: | Talaj, J.A, Bujacz, A, Bujacz, G. | | Deposit date: | 2018-09-13 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins.
Int J Mol Sci, 24, 2023
|
|
7BU0
 
 | |
5MUR
 
 | | X-ray structure of the F14'A mutant of GLIC in complex with propofol | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Sauguet, L, Fourati, Z, Delarue, M. | | Deposit date: | 2017-01-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for a Bimodal Allosteric Mechanism of General Anesthetic Modulation in Pentameric Ligand-Gated Ion Channels.
Cell Rep, 23, 2018
|
|
5IU4
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with ZM241385 at 1.7A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Segala, E, Guo, D, Cheng, R.K.Y, Bortolato, A, Deflorian, F, Dore, A.S, Errey, J.C, Heitman, L.H, Ijzerman, A.P, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength.
J.Med.Chem., 59, 2016
|
|
1LHV
 
 | | CRYSTAL STRUCTURE OF THE N-TERMINAL LG-DOMAIN OF SHBG IN COMPLEX WITH NORGESTREL | | Descriptor: | 13-BETA-ETHYL-17-ALPHA-ETHYNYL-17-BETA-HYDROXYGON-4-EN-3-ONE, CALCIUM ION, ISOPROPYL ALCOHOL, ... | | Authors: | Grishkovskaya, I, Avvakumov, G.V, Hammond, G.L, Catalano, M.G, Muller, Y.A. | | Deposit date: | 2002-04-17 | | Release date: | 2002-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Steroid Ligands Bind Human Sex Hormone-binding Globulin in Specific Orientations and Produce Distinct Changes in Protein Conformation
J.Biol.Chem., 277, 2002
|
|
1LSV
 
 | | Crystal structure of the CO-bound BjFixL heme domain | | Descriptor: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, Sensor protein FixL | | Authors: | Hao, B, Isaza, C, Arndt, J, Soltis, M, Chan, M.K. | | Deposit date: | 2002-05-19 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based mechanism of O2 sensing and ligand discrimination by
the FixL heme domain of Bradyrhizobium japonicum
Biochemistry, 41, 2002
|
|
6W3B
 
 | | Structure of apo unphosphorylated IRE1 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H, Mortara, K, Ferri, E, Rudolph, J, Wang, W. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
1LOX
 
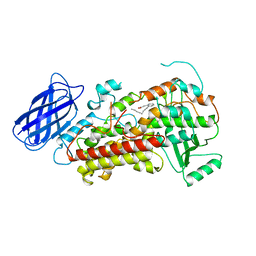 | | RABBIT RETICULOCYTE 15-LIPOXYGENASE | | Descriptor: | (2E)-3-(2-OCT-1-YN-1-YLPHENYL)ACRYLIC ACID, 15-LIPOXYGENASE, FE (II) ION | | Authors: | Gillmor, S.A, Villasenor, A, Fletterick, R.J, Sigal, E, Browner, M.F. | | Deposit date: | 1997-10-06 | | Release date: | 1998-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity.
Nat.Struct.Biol., 4, 1997
|
|
5AMI
 
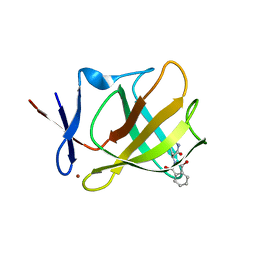 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, Wash I structure | | Descriptor: | CEREBLON ISOFORM 4, S-Thalidomide, ZINC ION | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
6OVP
 
 | |
1GMM
 
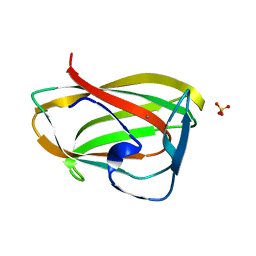 | | Carbohydrate binding module CBM6 from xylanase U Clostridium thermocellum | | Descriptor: | CALCIUM ION, CBM6, SODIUM ION, ... | | Authors: | Czjzek, M, Mosbah, A, Bolam, D, Allouch, J, Zamboni, V, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2001-09-19 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Location of the Ligand-Binding Site of Carbohydrate-Binding Modules that Have Evolved from a Common Sequence is not Conserved.
J.Biol.Chem., 276, 2001
|
|
1GHQ
 
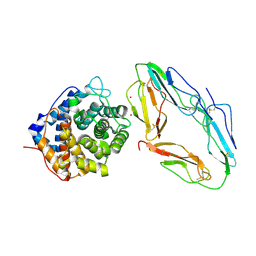 | | CR2-C3D COMPLEX STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, COMPLEMENT C3, CR2/CD121/C3D/EPSTEIN-BARR VIRUS RECEPTOR, ... | | Authors: | Szakonyi, G, Guthridge, J.M, Li, D, Holers, V.M, Chen, X.S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of complement receptor 2 in complex with its C3d ligand.
Science, 292, 2001
|
|
6A05
 
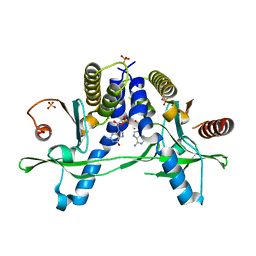 | | Structure of pSTING complex | | Descriptor: | 2-amino-9-[(2R,3R,3aR,5S,7aS,9R,10R,10aR,12R,14aS)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
7BDN
 
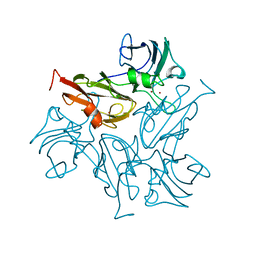 | | Structure of the Streptomyces coelicolor small laccase - cubic crystal form | | Descriptor: | COPPER (II) ION, Putative copper oxidase | | Authors: | Zovo, K, Majumdar, S, Lukk, T. | | Deposit date: | 2020-12-22 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substitution of the Methionine Axial Ligand of the T1 Copper for the Fungal-like Phenylalanine Ligand (M298F) Causes Local Structural Perturbations that Lead to Thermal Instability and Reduced Catalytic Efficiency of the Small Laccase from Streptomyces coelicolor A3(2).
Acs Omega, 7, 2022
|
|
5IU7
 
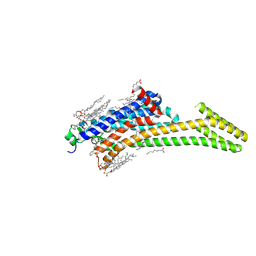 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with compound 12c at 1.9A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(furan-2-yl)-N~5~-[2-(4-phenylpiperidin-1-yl)ethyl][1,2,4]triazolo[1,5-a][1,3,5]triazine-5,7-diamine, ... | | Authors: | Segala, E, Guo, D, Cheng, R.K.Y, Bortolato, A, Deflorian, F, Dore, A.S, Errey, J.C, Heitman, L.H, Ijzerman, A.P, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength.
J.Med.Chem., 59, 2016
|
|
1PW2
 
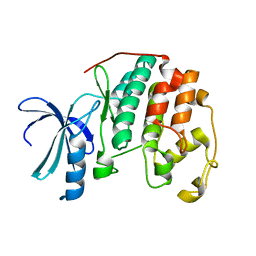 | | APO STRUCTURE OF HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-06-30 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the
activation loop.
Structure, 11, 2003
|
|
1PZ4
 
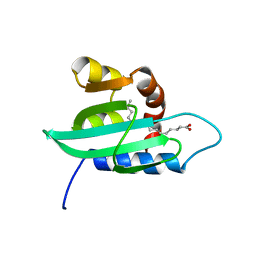 | | The structural determination of an insect (mosquito) Sterol Carrier Protein-2 with a ligand bound C16 Fatty Acid at 1.35 A resolution | | Descriptor: | PALMITIC ACID, sterol carrier protein 2 | | Authors: | Dyer, D.H, Lovell, S, Thoden, J.B, Holden, H.M, Rayment, I, Lan, Q. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Structural Determination of an Insect Sterol Carrier Protein-2 with a Ligand-bound C16 Fatty Acid at 1.35A Resolution
J.Biol.Chem., 278, 2003
|
|
5AMH
 
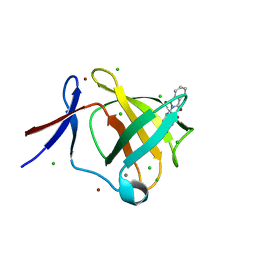 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, trigonal crystal form | | Descriptor: | CALCIUM ION, CEREBLON ISOFORM 4, CHLORIDE ION, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
4N6O
 
 | | Crystal structure of reduced legumain in complex with cystatin E/M | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IODIDE ION, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2013-10-14 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of an aspartimide-dependent Peptide ligase in human legumain.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3OB7
 
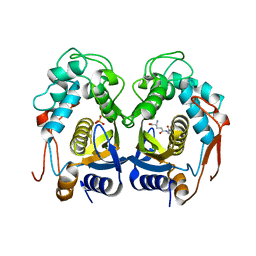 | | Human Thymidylate Synthase R163K with Cys 195 covalently modified by Glutathione | | Descriptor: | GLUTATHIONE, PHOSPHATE ION, Thymidylate synthase | | Authors: | Gibson, L.M, Celeste, L.R, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2010-08-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of human thymidylate synthase R163K with dUMP, FdUMP and glutathione show asymmetric ligand binding.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2GN1
 
 | |
3WVE
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, before photo-activation | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-05-17 | | Release date: | 2015-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3X26
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 5 min | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-10 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
