3G86
 
 | | Hepatitis C virus polymerase NS5B (BK 1-570) with thiazine inhibitor | | Descriptor: | N-{3-[6-fluoro-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl]-1,1-dioxido-4H-1,4-benzothiazin-7-yl}methanesulfonamide, NICKEL (II) ION, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-02-11 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 2: Synthesis and structure-activity relationships of benzothiazine-substituted quinolinediones
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3GGA
 
 | | HIV Protease inhibitors with pseudo-symmetric cores | | Descriptor: | V-1 protease, methyl [(1S,4S,5S,7S,10S)-4-benzyl-1,10-di-tert-butyl-5-hydroxy-2,9,12-trioxo-7-(4-pyridin-2-ylbenzyl)-13-oxa-3,8,11-triazatetradec-1-yl]carbamate | | Authors: | Stoll, V.S. | | Deposit date: | 2009-02-27 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus
protease inhibitors (A-792611 and A-790742) with potential for convenient
dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
2F46
 
 | |
3EIZ
 
 | |
2FAV
 
 | |
2FEA
 
 | |
2DVN
 
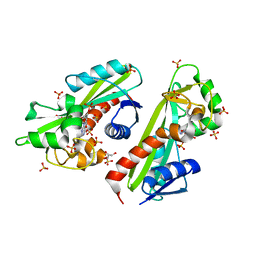 | |
1QHF
 
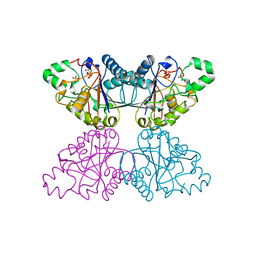 | | YEAST PHOSPHOGLYCERATE MUTASE-3PG COMPLEX STRUCTURE TO 1.7 A | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, PROTEIN (PHOSPHOGLYCERATE MUTASE), SULFATE ION | | Authors: | Crowhurst, G, Littlechild, J, Watson, H.C. | | Deposit date: | 1999-05-13 | | Release date: | 1999-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a phosphoglycerate mutase:3-phosphoglyceric acid complex at 1.7 A.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2F0C
 
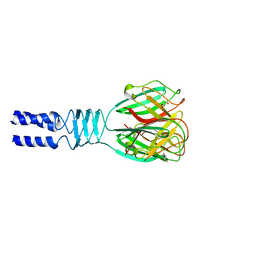 | |
1QTY
 
 | |
3EZO
 
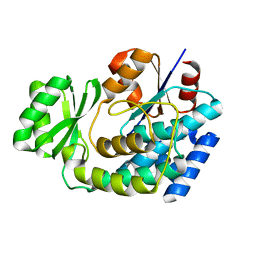 | |
1QMY
 
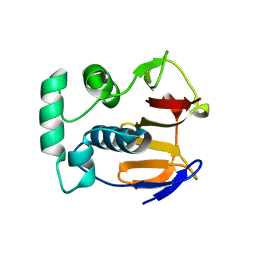 | | FMDV LEADER PROTEASE (LBSHORT-C51A-C133S) | | Descriptor: | 1,2-ETHANEDIOL, PROTEASE | | Authors: | Guarne, A, Tormo, J, Glaser, W, Skern, T, Fita, I. | | Deposit date: | 1999-10-08 | | Release date: | 2000-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Biochemical Features Distinguish the Foot-and-Mouth Disease Virus Leader Proteinase from Other Papain-Like Enzymes
J.Mol.Biol., 302, 2000
|
|
3GVF
 
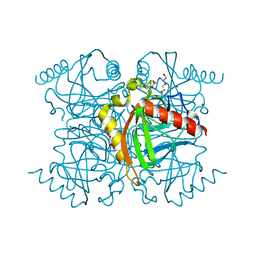 | |
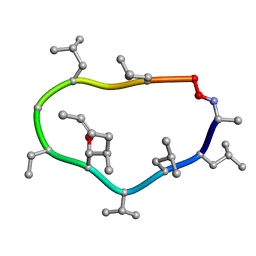
1CYB
 
 | |
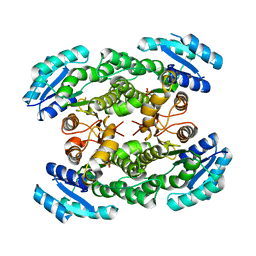
3GK3
 
 | |
2F05
 
 | |
2FSD
 
 | |
2J3S
 
 | |
2LL5
 
 | | Cyclo-TC1 Trp-cage | | Descriptor: | Cyclo-TC1 | | Authors: | Lin, J.C, Scian, M, Andersen, N.H. | | Deposit date: | 2011-10-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Crystal and NMR structures of a Trp-cage mini-protein benchmark for computational fold prediction.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6AKR
 
 | | Crystal structure of the PDE4D catalytic domain in complex with osthole | | Descriptor: | 7-methoxy-8-(3-methylbut-2-enyl)chromen-2-one, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Wang, S, Huo, Y.W, Xie, Y. | | Deposit date: | 2018-09-03 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.326 Å) | | Cite: | Airway relaxation mechanisms and structural basis of osthole for improving lung function in asthma.
Sci.Signal., 13, 2020
|
|
3L6V
 
 | | Crystal Structure of the Xanthomonas campestris Gyrase A C-terminal Domain | | Descriptor: | DNA gyrase subunit A | | Authors: | Hsieh, T.J, Yen, T.J, Lin, T.S, Chang, H.T, Huang, S.Y, Farh, L, Chan, N.L. | | Deposit date: | 2009-12-26 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Twisting of the DNA binding surface by a beta-strand-bearing proline modulates DNA gyrase activity
To be Published
|
|
2JAO
 
 | | Crystal structure of D12N variant of mouse cytosolic 5'(3')- deoxyribonucleotidase (cdN) in complex with deoxyguanosine 5'- monophosphate | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, 5'(3')-DEOXYRIBONUCLEOTIDASE, GLYCEROL, ... | | Authors: | Wallden, K, Ruzzenente, B, Bianchi, V, Nordlund, P. | | Deposit date: | 2006-11-29 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Human and Murine Deoxyribonucleotidases: Insights Into Recognition of Substrates and Nucleotide Analogues.
Biochemistry, 46, 2007
|
|
2ICC
 
 | | Extracellular Domain of CRIg | | Descriptor: | V-set and immunoglobulin domain-containing protein 4 | | Authors: | Wiesmann, C. | | Deposit date: | 2006-09-12 | | Release date: | 2006-11-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of C3b in complex with CRIg gives insights into regulation of complement activation.
Nature, 444, 2006
|
|
2ILT
 
 | | Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) with NADP and Adamantane Sulfone Inhibitor | | Descriptor: | 2-(2-CHLORO-4-FLUOROPHENOXY)-2-METHYL-N-[(1R,2S,3S,5S,7S)-5-(METHYLSULFONYL)-2-ADAMANTYL]PROPANAMIDE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Longenecker, K.L, Sorensen, B, Judge, R, Qin, W, Link, J.T. | | Deposit date: | 2006-10-03 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adamantane sulfone and sulfonamide 11-beta-HSD1 Inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3HD2
 
 | | Crystal structure of E. coli HPPK(Q50A) in complex with MgAMPCPP and pterin | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
