1T64
 
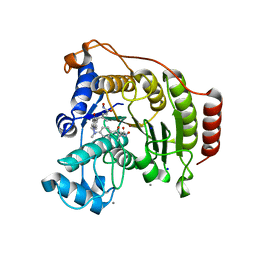 | | Crystal Structure of human HDAC8 complexed with Trichostatin A | | Descriptor: | CALCIUM ION, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
4O1G
 
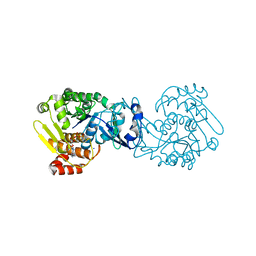 | | MTB adenosine kinase in complex with gamma-Thio-ATP | | Descriptor: | Adenosine kinase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SODIUM ION | | Authors: | Dostal, J, Brynda, J, Hocek, M, Pichova, I. | | Deposit date: | 2013-12-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
3PPX
 
 | | Crystal structure of the N1602A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
4O4X
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) double mutant Tyr-167-Ala and Trp-176-Ala from Haemophilus parasuis Hp5 | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-19 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4AYQ
 
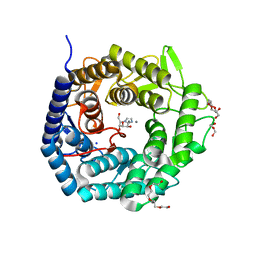 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4NLO
 
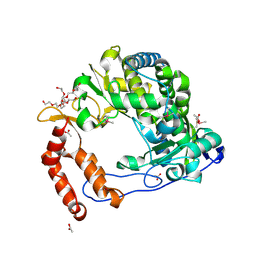 | | Poliovirus Polymerase - C290I Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4B5O
 
 | |
3F8V
 
 | |
3PKT
 
 | | Urate oxidase under 2 MPa / 20 bars pressure of nitrous oxide | | Descriptor: | 8-AZAXANTHINE, NITROUS OXIDE, SODIUM ION, ... | | Authors: | Marassio, G, Colloc'h, N, Prange, T, Abraini, J.H. | | Deposit date: | 2010-11-12 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: a crystallographic study.
Faseb J., 25, 2011
|
|
3PO4
 
 | | Structure of a mutant of the large fragment of DNA polymerase I from Thermus aquaticus in complex with a blunt-ended DNA and ddATP | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(2DA))-3'), DNA (5'-D(*TP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Learning from Directed Evolution: Thermus aquaticus DNA Polymerase Mutants with Translesion Synthesis Activity.
Chembiochem, 12, 2011
|
|
3Q3U
 
 | | Trametes cervina lignin peroxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Calvino, F.R, Romero, A, Miki, Y, Martinez, A.T. | | Deposit date: | 2010-12-22 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic, kinetic, and spectroscopic study of the first ligninolytic peroxidase presenting a catalytic tyrosine.
J.Biol.Chem., 286, 2011
|
|
4BRE
 
 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with transition state mimic adenosine 5'phosphovanadate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-PHOSPHOVANADATE, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
2V35
 
 | | Porcine Pancreatic Elastase in complex with inhibitor JM54 | | Descriptor: | (2R)-3-{[(BENZYLAMINO)CARBONYL]AMINO}-2-HYDROXYPROPANOIC ACID, ELASTASE-1, GLYCEROL, ... | | Authors: | Oliveira, T.F, Mulchande, J, Martins, L, Moreira, R, Iley, J, Archer, M. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Efficiency of C-4 Substituents in Activating the Beta-Lactam Scaffold Towards Serine Proteases and Hydroxide Ion.
Org.Biomol.Chem., 5, 2007
|
|
3FOB
 
 | | Crystal structure of bromoperoxidase from Bacillus anthracis | | Descriptor: | Bromoperoxidase, CHLORIDE ION, SODIUM ION | | Authors: | Osipiuk, J, Gu, M, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | X-ray crystal structure of bromoperoxidase from Bacillus anthracis.
To be Published
|
|
4NW7
 
 | | PDE4 catalytic domain | | Descriptor: | (4-{[4-(3-chlorophenyl)-6-cyclopropyl-1,3,5-triazin-2-yl]amino}phenyl)acetic acid, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Fox III, D, Edwards, T.E. | | Deposit date: | 2013-12-05 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of triazines as selective PDE4B versus PDE4D inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3FKR
 
 | | Structure of L-2-keto-3-deoxyarabonate dehydratase complex with pyruvate | | Descriptor: | L-2-keto-3-deoxyarabonate dehydratase, PHOSPHATE ION, SODIUM ION | | Authors: | Shimada, N, Mikami, B. | | Deposit date: | 2008-12-17 | | Release date: | 2010-02-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural analysis of L -2-keto-3-deoxyarabonate dehydratase an enzyme involved in an alternative bacterial pathway of L-arabinose metabolism in complex with pyruvate
To be Published
|
|
2V5C
 
 | | Family 84 glycoside hydrolase from Clostridium perfringens, 2.1 Angstrom structure | | Descriptor: | CACODYLATE ION, CALCIUM ION, O-GLCNACASE NAGJ, ... | | Authors: | Ficko-Blean, E, Gregg, K.J, Adams, J.J, Hehemann, J.H, Smith, S.J, Czjzek, M, Boraston, A.B. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Portrait of an Enzyme: A Complete Structural Analysis of a Multi-Modular Beta-N-Acetylglucosaminidase from Clostridium Perfringens
J.Biol.Chem., 284, 2009
|
|
4F06
 
 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 RPB_2270 in complex with P-HYDROXYBENZOIC ACID | | Descriptor: | Extracellular ligand-binding receptor, GLYCEROL, P-HYDROXYBENZOIC ACID, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-15 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
4F6U
 
 | | Crystal structure of human CDK8/CYCC in complex with compound 5 (1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[3-(morpholin-4-yl)propyl]urea) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[3-(morpholin-4-yl)propyl]urea, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4O34
 
 | | Room temperature macromolecular serial crystallography using synchrotron radiation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Stellato, F, Oberthuer, D, Liang, M, Bean, R, Gati, C, Yefanov, O, Barty, A, Burkhardt, A, Fischer, P, Galli, L, Kirian, R.A, Mayer, J, Pannerselvam, S, Yoon, C.H, Chervinskii, F, Speller, E, White, T.A, Betzel, C, Meents, A, Chapman, H.N. | | Deposit date: | 2013-12-18 | | Release date: | 2014-06-11 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Room-temperature macromolecular serial crystallography using synchrotron radiation.
IUCrJ, 1, 2014
|
|
2XH8
 
 | | X-ray structure of 119-141 ZnuA deletion mutant from Salmonella enterica. | | Descriptor: | SODIUM ION, SULFATE ION, ZINC ABC TRANSPORTER, ... | | Authors: | Alaleona, F, Ilari, A, Battistoni, A, Petrarca, P, Chiancone, E. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The X-Ray Structure of the Zinc Transporter Znua from Salmonella Enterica Discloses a Unique Triad of Zinc Coordinating Histidines.
J.Mol.Biol., 409, 2011
|
|
3GZA
 
 | |
2XN4
 
 | | Crystal structure of the kelch domain of human KLHL2 (Mayven) | | Descriptor: | 1,2-ETHANEDIOL, KELCH-LIKE PROTEIN 2, SODIUM ION, ... | | Authors: | Canning, P, Hozjan, V, Cooper, C.D.O, Ayinampudi, V, Vollmar, M, Pike, A.C.W, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2010-07-30 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
2XXU
 
 | | Crystal structure of the GluK2 (GluR6) M770K LBD dimer in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
4O57
 
 | | Crystal Structure of Trichomonas vaginalis Triosephosphate Isomerase Ile45-Tyr mutant (Tvag_497370) | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Lara-Gonzalez, S, Estrella-Hernandez, P, Montero-Moran, G.M, Benitez-Cardoza, C.G, Brieba, L.G. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-24 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Engineering mutants with altered dimer-monomer equilibrium reveal the existence of stable monomeric Triosephosphate isomerases
To be Published
|
|
