8SXY
 
 | | X-ray crystal structure of UDP- 2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27 in complex with its product UDP-2,3-diacetamido-2,3-dideoxy-d-mannuronic acid at pH 5 | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, CHLORIDE ION, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase | | Authors: | McKnight, J.O, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
8SY9
 
 | | X-ray crystal structure of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, D98N variant in the presence of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid, UDP-N-acetylglucosamine and UDP at pH 7 | | Descriptor: | (2~{S},3~{S},4~{R},5~{R},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
8SYD
 
 | | X-ray crystal structure of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, D98N variant in the presence of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid and UDP-N-acetylglucosamine at pH 6 | | Descriptor: | (2~{S},3~{S},4~{R},5~{R},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, CHLORIDE ION, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase, ... | | Authors: | Kroft, C.W, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
8SYE
 
 | | X-ray crystal structure of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, D98N variant in the presence of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid and UDP at pH 6 | | Descriptor: | (2~{S},3~{S},4~{R},5~{R},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, CHLORIDE ION, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase, ... | | Authors: | Jast, J.D.T, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
8SXV
 
 | | X-ray crystal structure of UDP- 2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, apo form, pH 9 | | Descriptor: | CHLORIDE ION, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase | | Authors: | McKnight, J.O, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
8SYH
 
 | | X-ray crystal structure of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, D98N variant in the presence of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid and UDP at pH 8 | | Descriptor: | (2~{S},3~{S},4~{R},5~{R},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, CHLORIDE ION, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase, ... | | Authors: | Jast, J.D.T, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
6DCF
 
 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and bound to kanglemycin A | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
8SYB
 
 | | X-ray crystal structure of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, D98N variant in the presence of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid and UDP-N-acetylglucosamine at pH 9 | | Descriptor: | (2~{S},3~{S},4~{R},5~{R},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, CHLORIDE ION, SODIUM ION, ... | | Authors: | Kroft, C.W, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
9BR7
 
 | | Crystal structure of human succinyl-CoA:glutarate-CoA transferase (SUGCT) in complex with Losartan carboxylic acid | | Descriptor: | AMMONIUM ION, SULFATE ION, Succinate--hydroxymethylglutarate CoA-transferase, ... | | Authors: | Wu, R, Lazarus, M.B. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization, Structure, and Inhibition of the Human Succinyl-CoA:glutarate-CoA Transferase, a Putative Genetic Modifier of Glutaric Aciduria Type 1.
Acs Chem.Biol., 19, 2024
|
|
6DCY
 
 | |
9ATK
 
 | |
9ASX
 
 | |
9ATU
 
 | |
7BHD
 
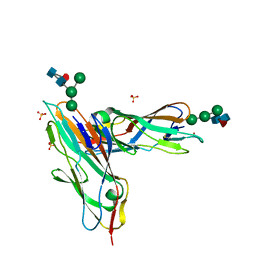 | | FimH in complex with alpha1,6 core-fucosylated oligomannose-3, crystallized in the trigonal space group | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bridot, C, Bouckaert, J, Krammer, E.-M. | | Deposit date: | 2021-01-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
2LK0
 
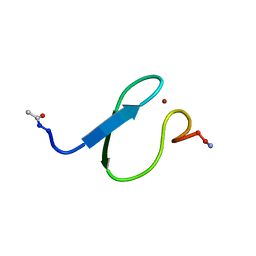 | |
6D9W
 
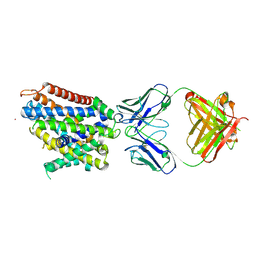 | | Crystal structure of Deinococcus radiodurans MntH, an Nramp-family transition metal transporter, in the inward-open apo state | | Descriptor: | Divalent metal cation transporter MntH, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Gaudet, R, Bane, L.B, Weihofen, W.A, Singharoy, A, Zimanyi, C.M, Bozzi, A.T. | | Deposit date: | 2018-04-30 | | Release date: | 2019-02-13 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (3.941 Å) | | Cite: | Structures in multiple conformations reveal distinct transition metal and proton pathways in an Nramp transporter.
Elife, 8, 2019
|
|
2L02
 
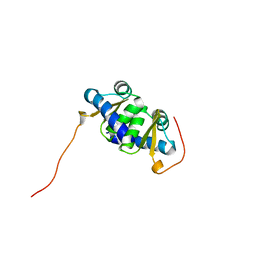 | | Solution NMR Structure of protein BT2368 from Bacteroides thetaiotaomicron, Northeast Structural Genomics Consortium Target BtR375 | | Descriptor: | Uncharacterized protein | | Authors: | Eletsky, A, Lee, H, Wang, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-29 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of protein BT2368 from Bacteroides thetaiotaomicron
To be Published
|
|
2LGL
 
 | | NMR structure of the UHRF1 PHD domain | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
6DI3
 
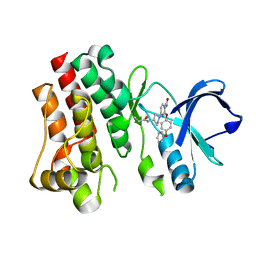 | | CRYSTAL STRUCTURE OF BTK IN COMPLEX WITH FRAGMENT LIGAND | | Descriptor: | 6-[(3S)-3-(acryloylamino)pyrrolidin-1-yl]-2-(4-phenoxyphenoxy)pyridine-3-carboxamide, Tyrosine-protein kinase BTK | | Authors: | GARDBERG, A. | | Deposit date: | 2018-05-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of potent, highly selective covalent irreversible BTK inhibitors from a fragment hit.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
2LLP
 
 | | Solution structure of a THP type 1 alpha 1 collagen fragment (772-786) | | Descriptor: | Collagen alpha-1(I) chain | | Authors: | Bertini, I, Fragai, M, Luchinat, C, Melikian, M, Toccafondi, M, Lauer, J.L, Fields, G.B, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for matrix metalloproteinase 1-catalyzed collagenolysis.
J.Am.Chem.Soc., 134, 2012
|
|
6DAN
 
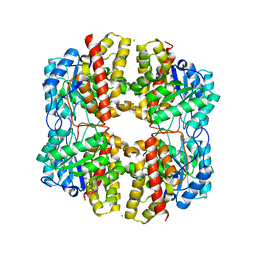 | | PhdJ WT 2 Angstroms resolution | | Descriptor: | CHLORIDE ION, PhdJ | | Authors: | Medellin, B.P, LeVieux, J.A, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural Characterization of the Hydratase-Aldolases, NahE and PhdJ: Implications for the Specificity, Catalysis, and N-Acetylneuraminate Lyase Subgroup of the Aldolase Superfamily.
Biochemistry, 57, 2018
|
|
2M3U
 
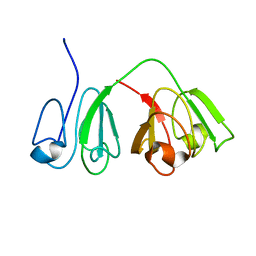 | |
2M5P
 
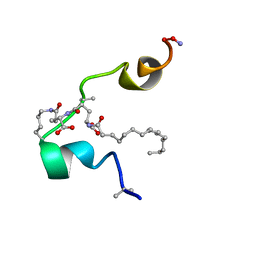 | |
2M9P
 
 | |
8SVC
 
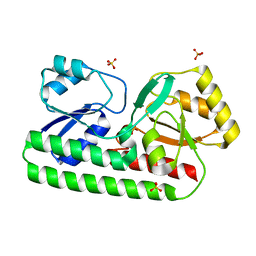 | | Crystal Structure of SBP from Klebsiella pneumoniae | | Descriptor: | Metal ABC transporter substrate-binding protein, SULFATE ION, ZINC ION | | Authors: | Giles, M.W, Cole, G.B, Ng, D, McDevitt, C.A, Moraes, T.F. | | Deposit date: | 2023-05-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Zinc acquisition and its contribution to Klebsiella pneumoniae virulence.
Front Cell Infect Microbiol, 13, 2023
|
|
