1K0J
 
 | | Pseudomonas aeruginosa phbh R220Q in complex with NADPH and free of p-OHB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, P-HYDROXYBENZOATE HYDROXYLASE, ... | | Authors: | Wang, J, Ortiz-Maldonado, M, Entsch, B, Ballou, D, Gatti, D.L. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K0I
 
 | | Pseudomonas aeruginosa phbh R220Q in complex with 100mM PHB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID, ... | | Authors: | Wang, J, Ortiz-Maldonado, M, Entsch, B, Ballou, D, Gatti, D.L. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K0L
 
 | | Pseudomonas aeruginosa phbh R220Q free of p-OHB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, SULFATE ION, ... | | Authors: | Wang, J, Ortiz-Maldonado, M, Entsch, B, Ballou, D, Gatti, D.L. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1B2R
 
 | | FERREDOXIN-NADP+ REDUCTASE (MUTATION: E 301 A) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN-NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-11-27 | | Release date: | 1999-12-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the catalytic role of Glu301 in Anabaena PCC 7119 ferredoxin-NADP+ reductase revealed by x-ray crystallography.
Proteins, 38, 2000
|
|
7Z4X
 
 | |
1BJK
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH ARG 264 REPLACED BY GLU (R264E) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Arg100 and Arg264 from Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal NADP+ binding and electron transfer.
Biochemistry, 37, 1998
|
|
1KRH
 
 | | X-ray Structure of Benzoate Dioxygenase Reductase | | Descriptor: | Benzoate 1,2-Dioxygenase Reductase, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Karlsson, A, Beharry, Z.M, Eby, D.M, Coulter, E.D, Niedle, E.L, Kurtz Jr, D.M, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of benzoate 1,2-dioxygenase reductase from Acinetobacter sp. strain ADP1.
J.Mol.Biol., 318, 2002
|
|
2HJ3
 
 | | Structure of the Arabidopsis Thaliana Erv1 Thiol Oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Sulfhydryl oxidase Erv1p | | Authors: | Vitu, E, Fass, D. | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Gain of Function in an ERV/ALR Sulfhydryl Oxidase by Molecular Engineering of the Shuttle Disulfide.
J.Mol.Biol., 362, 2006
|
|
7C3L
 
 | | Structure of L-lysine oxidase D212A/D315A in complex with L-tyrosine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, L-lysine oxidase, ... | | Authors: | Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
7C3J
 
 | | Structure of L-lysine oxidase D212A/D315A in complex with L-phenylalanine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, L-lysine oxidase, ... | | Authors: | Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
3HDQ
 
 | |
3HE3
 
 | |
6WPU
 
 | |
2YG5
 
 | | Structure-based redesign of cofactor binding in Putrescine Oxidase: A394C mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PUTRESCINE OXIDASE, ... | | Authors: | Kopacz, M.M, Rovida, S, van Duijn, E, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Redesign of Cofactor Binding in Putrescine Oxidase.
Biochemistry, 50, 2011
|
|
7MWT
 
 | |
8JDS
 
 | | Crystal structure of mLDHD in complex with Pyruvate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MANGANESE (II) ION, PYRUVIC ACID, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.636 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDC
 
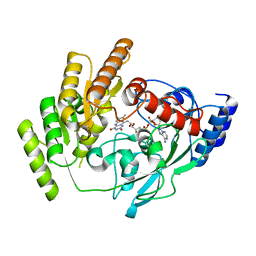 | | Crystal structure of mLDHD in apo form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, mitochondrial | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDD
 
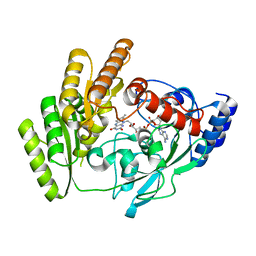 | | Crystal structure of H405A mLDHD in apo form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, mitochondrial | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDF
 
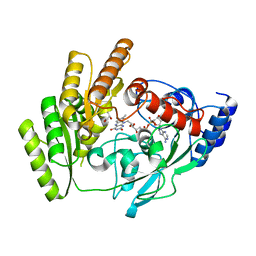 | | Crystal structure of H405A mLDHD in complex with D-lactate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LACTIC ACID, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDE
 
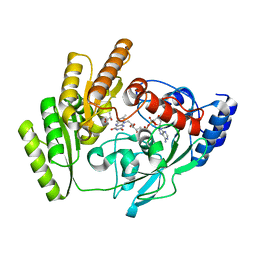 | | Crystal structure of mLDHD in complex with D-lactate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LACTIC ACID, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDP
 
 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxyisovaleric acid | | Descriptor: | DEAMINOHYDROXYVALINE, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
1TYT
 
 | |
1MOK
 
 | | NADPH DEPENDENT 2-KETOPROPYL COENZYME M OXIDOREDUCTASE/CARBOXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, orf3 | | Authors: | Nocek, B, Jang, S.B, Jeong, M.S, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2002-09-09 | | Release date: | 2002-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for CO2 Fixation by a Novel Member of the Disulfide Oxidoreductase Family of Enzymes,
2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase
Biochemistry, 41, 2002
|
|
1FDR
 
 | | FLAVODOXIN REDUCTASE FROM E. COLI | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVODOXIN REDUCTASE | | Authors: | Ingelman, M, Bianchi, V, Eklund, H. | | Deposit date: | 1997-03-06 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The three-dimensional structure of flavodoxin reductase from Escherichia coli at 1.7 A resolution.
J.Mol.Biol., 268, 1997
|
|
1TIW
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with L-Tetrahydro-2-furoic acid | | Descriptor: | Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE, TETRAHYDROFURAN-2-CARBOXYLIC ACID | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-02 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
