5MMW
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 6 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[3-(2-methyl-3-phenyl-phenyl)-4-oxidanyl-phenyl]prop-2-enoic acid, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-12 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
1DP0
 
 | | E. COLI BETA-GALACTOSIDASE AT 1.7 ANGSTROM | | Descriptor: | BETA-GALACTOSIDASE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 1999-12-22 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
3OD2
 
 | | E. coli NikR soaked with excess nickel ions | | Descriptor: | 3-CYCLOHEXYLPROPYL 4-O-ALPHA-D-GLUCOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, NICKEL (II) ION, Nickel-responsive regulatory protein | | Authors: | Phillips, C.M, Schreiter, E.R, Drennan, C.L. | | Deposit date: | 2010-08-10 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of low-affinity nickel binding to the nickel-responsive transcription factor NikR from Escherichia coli.
Biochemistry, 49, 2010
|
|
3OSR
 
 | | Maltose-bound maltose sensor engineered by insertion of circularly permuted green fluorescent protein into E. coli maltose binding protein at position 311 | | Descriptor: | Maltose-binding periplasmic protein,Green fluorescent protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Echevarria, I.M, Marvin, J.S, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2010-09-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A genetically encoded, high-signal-to-noise maltose sensor.
Proteins, 79, 2011
|
|
3OSQ
 
 | | Maltose-bound maltose sensor engineered by insertion of circularly permuted green fluorescent protein into E. coli maltose binding protein at position 175 | | Descriptor: | Maltose-binding periplasmic protein,Green fluorescent protein, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Echevarria, I.M, Marvin, J.S, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2010-09-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A genetically encoded, high-signal-to-noise maltose sensor.
Proteins, 79, 2011
|
|
8DBR
 
 | | E. coli ATP synthase imaged in 10mM MgATP State2 "half-up | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
8DBT
 
 | | E. coli ATP synthase imaged in 10mM MgATP State2 "down | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
8DBP
 
 | | E. coli ATP synthase imaged in 10mM MgATP State1 "half-up | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
8DBQ
 
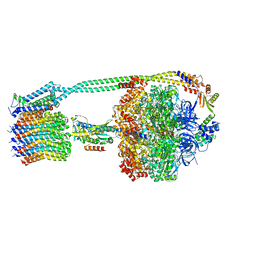 | |
8DBV
 
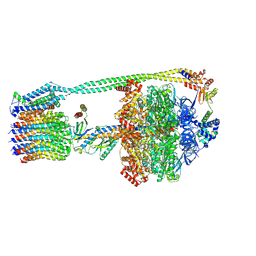 | | E. coli ATP synthase imaged in 10mM MgATP State3 "down | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
8DBS
 
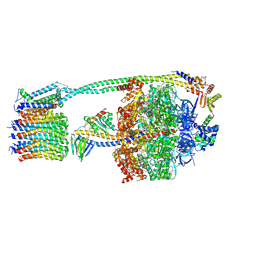 | |
8DBU
 
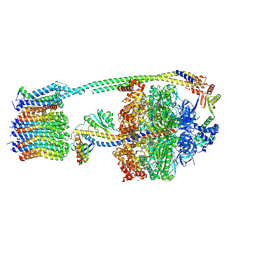 | |
8DBW
 
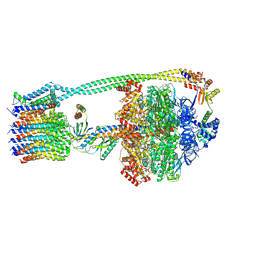 | |
2XX4
 
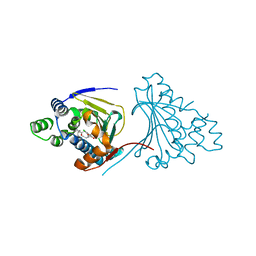 | | Macrolactone Inhibitor bound to HSP90 N-term | | Descriptor: | (E)-ETHYL 13-CHLORO-14,16-DIHYDROXY-1,11-DIOXO-1,2,3,4,7,8,9,10,11,12-DECAHYDROBENZO[C][1]AZACYCLOTETRADECINE-10-CARBOXYLATE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Moody, C.J, Prodromou, C, Pearl, L.H, Roe, S.M. | | Deposit date: | 2010-11-08 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Targeting the Hsp90 Molecular Chaperone with Novel Macrolactams. Synthesis, Structural, Binding, and Cellular Studies.
Acs Chem.Biol., 6, 2011
|
|
8F6C
 
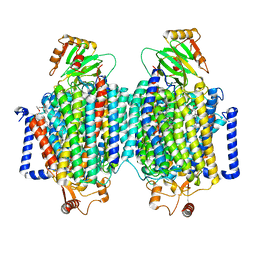 | | E. coli cytochrome bo3 ubiquinol oxidase dimer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Guo, Y, Karimullina, E, Borek, D, Savchenko, A. | | Deposit date: | 2022-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Monomer and dimer structures of cytochrome bo 3 ubiquinol oxidase from Escherichia coli.
Protein Sci., 32, 2023
|
|
8F68
 
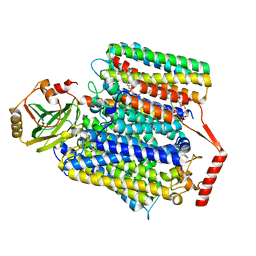 | | E. coli cytochrome bo3 ubiquinol oxidase monomer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Guo, Y, Karimullina, E, Borek, D, Savchenko, A. | | Deposit date: | 2022-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Monomer and dimer structures of cytochrome bo 3 ubiquinol oxidase from Escherichia coli.
Protein Sci., 32, 2023
|
|
8SXB
 
 | |
1F4H
 
 | | E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1F4A
 
 | | E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
5LRY
 
 | | E coli [NiFe] Hydrogenase Hyd-1 mutant E28D | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Evans, R.M, Brooke, E.J, Armstrong, F.A. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J.Am.Chem.Soc., 140, 2018
|
|
6QEM
 
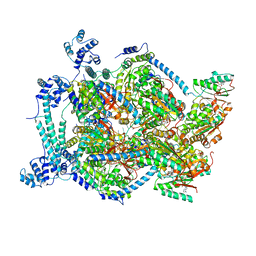 | | E. coli DnaBC complex bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication protein DnaC, MAGNESIUM ION, ... | | Authors: | Arias-Palomo, E, Puri, N, O'Shea Murray, V.L, Yan, Q, Berger, J.M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Physical Basis for the Loading of a Bacterial Replicative Helicase onto DNA.
Mol.Cell, 74, 2019
|
|
6S1K
 
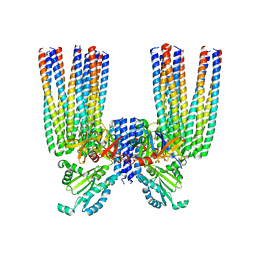 | | E. coli Core Signaling Unit, carrying QQQQ receptor mutation | | Descriptor: | CheW, Chemotaxis protein CheA, Methyl-accepting chemotaxis protein I | | Authors: | Cassidy, C.K. | | Deposit date: | 2019-06-18 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.38 Å) | | Cite: | Structure and dynamics of the E. coli chemotaxis core signaling complex by cryo-electron tomography and molecular simulations.
Commun Biol, 3, 2020
|
|
3I3B
 
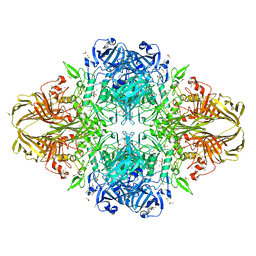 | | E.coli (lacz) Beta-Galactosidase (M542A) in Complex with D-Galactopyranosyl-1-on | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Dymianiw, D, Minhas, B, Huber, R.E. | | Deposit date: | 2009-06-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of Met-542 as a guide for the conformational changes of Phe-601 that occur during the reaction of β-galactosidase (Escherichia coli).
Biochem.Cell Biol., 88, 2010
|
|
3I3E
 
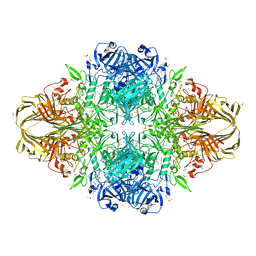 | | E. COLI (lacZ) BETA-GALACTOSIDASE (M542A) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Dugdale, M.L, Dymianiw, D, Minhas, B, Huber, R.E. | | Deposit date: | 2009-06-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of Met-542 as a guide for the conformational changes of Phe-601 that occur during the reaction of β-galactosidase (Escherichia coli).
Biochem.Cell Biol., 88, 2010
|
|
8Y7L
 
 | |
