4M61
 
 | |
6W35
 
 | |
2XQC
 
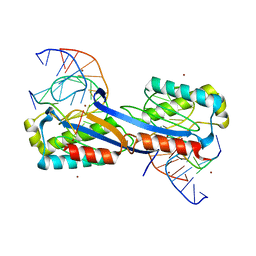 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE COMPLEXED WITH LEFT END RECOGNITION AND CLEAVAGE SITE AND ZN | | Descriptor: | 5'-D(TP*TP*GP*AP*TP*GP)-3', DRA2 TRANSPOSASE LEFT END RECOGNITION SEQUENCE, TRANSPOSASE, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-09-01 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
1I6J
 
 | |
1NAB
 
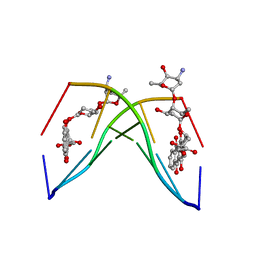 | | The crystal structure of the complex between a disaccharide anthracycline and the DNA hexamer d(CGATCG) reveals two different binding sites involving two DNA duplexes | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*G)-3', 7-[5-(4-AMINO-5-HYDROXY-6-METHYL-TETRAHYDRO-PYRAN-2-YLOXY)-4-HYDROXY-6-METHYL-TETRAHYDRO-PYRAN-2-YLOXY]-6,9,11-TRIHYDROXY-9-(2-HYDROXY-ACETYL)-7,8,9,10-TETRAHYDRO-NAPHTHACENE-5,12-DIONE | | Authors: | Temperini, C, Messori, L, Orioli, P, Di Bugno, C, Animati, F, Ughetto, G. | | Deposit date: | 2002-11-27 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the complex between a disaccharide anthracycline and the DNA hexamer d(CGATCG) reveals two different binding sites involving two DNA duplexes
Nucleic Acids Res., 31, 2003
|
|
5OL5
 
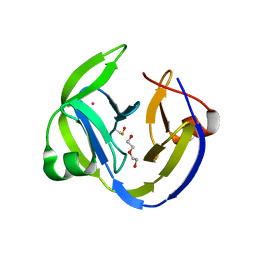 | | Crystal structure of an inactivated Ssp SICLOPPS intein with CFAHPQ extein | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, DNA polymerase III subunit alpha,DNA polymerase III subunit alpha, ... | | Authors: | Kick, L.M, Schneider, S. | | Deposit date: | 2017-07-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | Mechanistic Insights into Cyclic Peptide Generation by DnaE Split-Inteins through Quantitative and Structural Investigation.
Chembiochem, 18, 2017
|
|
5E5S
 
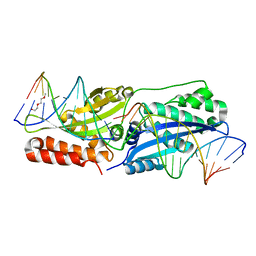 | | I-SmaMI K103A mutant | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Bottom strand DNA, DNA (5'-D(P*CP*AP*GP*GP*TP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Shen, B.W. | | Deposit date: | 2015-10-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Structural Basis of Asymmetry in DNA Binding and Cleavage as Exhibited by the I-SmaMI LAGLIDADG Meganuclease.
J.Mol.Biol., 428, 2016
|
|
3SSF
 
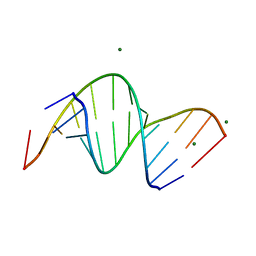 | | Crystal structure of RNA:DNA dodecamer corresponding to HIV-1 polypurine tract, at 1.6 A resolution. | | Descriptor: | 5'-D(*CP*CP*TP*TP*TP*TP*CP*TP*TP*TP*TP*A)-3', 5'-R(*UP*AP*AP*AP*AP*GP*AP*AP*AP*AP*GP*G)-3', MAGNESIUM ION | | Authors: | Drozdzal, P, Michalska, K, Kierzek, R, Lomozik, L, Jaskolski, M. | | Deposit date: | 2011-07-08 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of an RNA/DNA dodecamer corresponding to the HIV-1 polypurine tract at 1.6 Angstrom resolution
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1RSB
 
 | |
8VX9
 
 | |
2BAM
 
 | |
2XO6
 
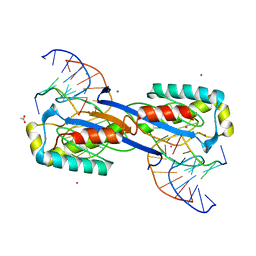 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE Y132F MUTANT COMPLEXED WITH LEFT END RECOGNITION AND CLEAVAGE SITE | | Descriptor: | 5'-D(*TP*TP*GP*AP*TP*G)-3', ACETATE ION, CADMIUM ION, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-08-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
4BG7
 
 | |
5TRD
 
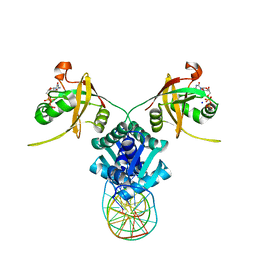 | | Structure of RbkR (Riboflavin Kinase) from Thermoplasma acidophilum determined in complex with CTP and its cognate DNA operator | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*TP*TP*AP*CP*TP*AP*AP*TP*TP*CP*AP*CP*GP*AP*GP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*AP*CP*TP*CP*GP*TP*GP*AP*AP*TP*TP*AP*GP*TP*AP*A)-3'), ... | | Authors: | Vetting, M.W, Rodionova, I.A, Li, X, Osterman, A.L, Rodionov, D.A, Almo, S.C. | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of RbkR (Riboflavin Kinase) from Thermoplasma acidophilum determined in complex with CTP and its cognate DNA operator
To be published
|
|
4HDV
 
 | |
2HXM
 
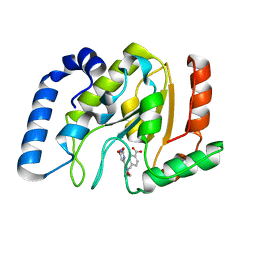 | | Complex of UNG2 and a small Molecule synthetic Inhibitor | | Descriptor: | 4-[(1E,7E)-8-(2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDIN-4-YL)-3,6-DIOXA-2,7-DIAZAOCTA-1,7-DIEN-1-YL]BENZOIC ACID, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Ghung, S, Seiple, L, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2006-08-03 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mimicking damaged DNA with a small molecule inhibitor of human UNG2.
Nucleic Acids Res., 34, 2006
|
|
4L6P
 
 | |
1Z1B
 
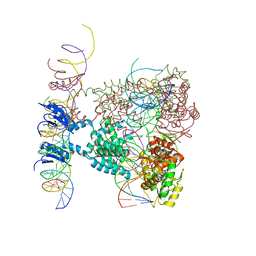 | | Crystal structure of a lambda integrase dimer bound to a COC' core site | | Descriptor: | 26-MER DNA, 29-MER DNA, 5'-D(*CP*T*CP*GP*TP*TP*CP*AP*GP*CP*TP*TP*TP*TP*TP*T)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
384D
 
 | |
2ZXO
 
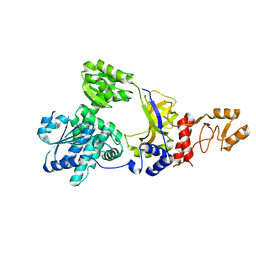 | | Crystal structure of RecJ from Thermus thermophilus HB8 | | Descriptor: | Single-stranded DNA specific exonuclease RecJ | | Authors: | Wakamatsu, T, Kitamura, Y, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of RecJ exonuclease defines its specificity for single-stranded DNA
J.Biol.Chem., 285, 2010
|
|
1RPZ
 
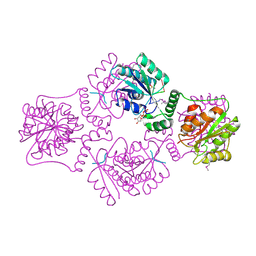 | | T4 POLYNUCLEOTIDE KINASE BOUND TO 5'-TGCAC-3' SSDNA | | Descriptor: | 5'-D(*TP*GP*CP*AP*C)-3', ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Eastberg, J.H, Pelletier, J, Stoddard, B.L. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase.
Nucleic Acids Res., 32, 2004
|
|
4HDU
 
 | |
2ZXP
 
 | | Crystal structure of RecJ in complex with Mn2+ from Thermus thermophilus HB8 | | Descriptor: | MANGANESE (II) ION, Single-stranded DNA specific exonuclease RecJ | | Authors: | Wakamatsu, T, Kitamura, Y, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of RecJ exonuclease defines its specificity for single-stranded DNA
J.Biol.Chem., 285, 2010
|
|
2ZXR
 
 | | Crystal structure of RecJ in complex with Mg2+ from Thermus thermophilus HB8 | | Descriptor: | MAGNESIUM ION, Single-stranded DNA specific exonuclease RecJ | | Authors: | Wakamatsu, T, Kitamura, Y, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of RecJ exonuclease defines its specificity for single-stranded DNA
J.Biol.Chem., 285, 2010
|
|
1RRC
 
 | | T4 POLYNUCLEOTIDE KINASE BOUND TO 5'-GTC-3' SSDNA | | Descriptor: | 5'-D(*GP*TP*C)-3', ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Eastberg, J.H, Pelletier, J, Stoddard, B.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase.
Nucleic Acids Res., 32, 2004
|
|
