4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
8HXZ
 
 | | Cryo-EM structure of Eaf3 CHD in complex with nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (352-MER), Histone H2A, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7SGL
 
 | | DNA-PK complex of DNA end processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, Hairpin_1, ... | | Authors: | Liu, L, Li, J, Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2021-10-06 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
1ID3
 
 | | CRYSTAL STRUCTURE OF THE YEAST NUCLEOSOME CORE PARTICLE REVEALS FUNDAMENTAL DIFFERENCES IN INTER-NUCLEOSOME INTERACTIONS | | Descriptor: | HISTONE H2A.1, HISTONE H2B.2, HISTONE H3, ... | | Authors: | White, C.L, Suto, R.K, Luger, K. | | Deposit date: | 2001-04-03 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions.
EMBO J., 20, 2001
|
|
3BHO
 
 | |
7P15
 
 | |
7OZW
 
 | |
6I7B
 
 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 3. | | Descriptor: | Nucleoprotein | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-11-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6TWU
 
 | | MAGI1_2 complexed with a phosphomimetic 16E6 peptide | | Descriptor: | CALCIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Structure, 28, 2020
|
|
6TWQ
 
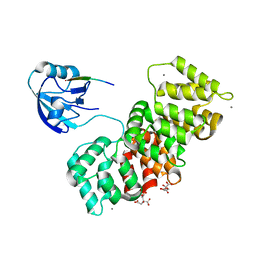 | | MAGI1_2 complexed with a 16E6 peptide | | Descriptor: | CALCIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Structure, 28, 2020
|
|
4TU0
 
 | | CRYSTAL STRUCTURE OF CHIKUNGUNYA VIRUS NSP3 MACRO DOMAIN IN COMPLEX WITH A 2'-5' OLIGOADENYLATE TRIMER | | Descriptor: | 2'-5' OLIGOADENYLATE TRIMER, DI(HYDROXYETHYL)ETHER, Non-structural polyprotein 3 | | Authors: | Morin, B, Ferron, f.p, Malet, h, Coutard, b, Canard, b. | | Deposit date: | 2014-06-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF CHIKUNGUNYA VIRUS NSP3 MACRO DOMAIN IN COMPLEX WITH A 2'-5' OLIGOADENYLATE TRIMER
TO BE PUBLISHED
|
|
6MSJ
 
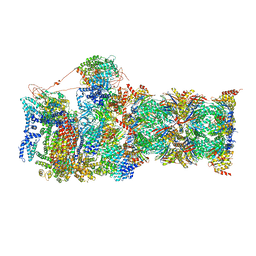 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSG
 
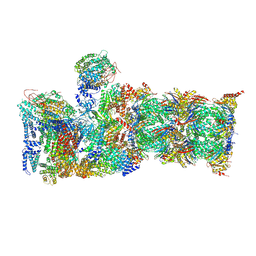 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSE
 
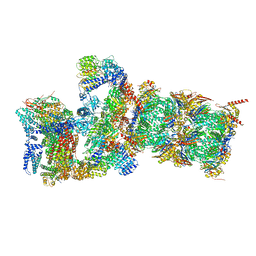 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSB
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSK
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSH
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSD
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6H9G
 
 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 1. | | Descriptor: | Nucleoprotein, Polypeptide loop | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Munier, S, Carlero, D, Naffakh, N, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-08-03 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6K15
 
 | | RSC substrate-recruitment module | | Descriptor: | Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
7ZI4
 
 | | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Vance, N.R, Ayala, R, Willhoft, O, Tvardovskiy, A, McCormack, E.A, Bartke, T, Zhang, X, Wigley, D.B. | | Deposit date: | 2022-04-07 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome
To Be Published
|
|
6I7M
 
 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 4. | | Descriptor: | Nucleoprotein | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-11-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
5WG6
 
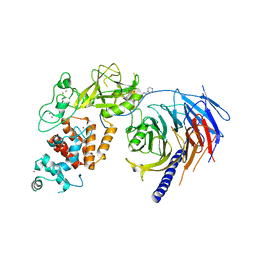 | | Human Polycomb Repressive Complex 2 in complex with GSK126 inhibitor | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine N-methyltransferase EZH2,Polycomb protein SUZ12 (E.C.2.1.1.43) chimera, Polycomb protein EED, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.901 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
7KSO
 
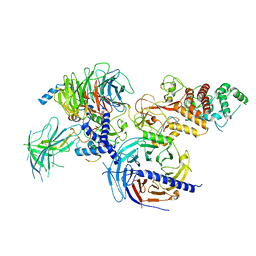 | | Cryo-EM structure of PRC2:EZH1-AEBP2-JARID2 | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
4RKW
 
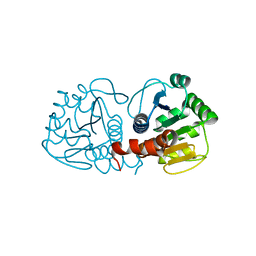 | | Crystal structure of DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Liddington, R.C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in parkinson's disease models.
J.Neurosci., 34, 2014
|
|
