6LDP
 
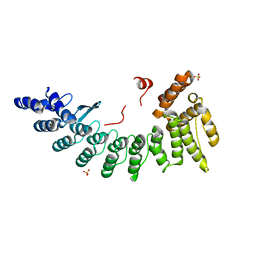 | | Structure of CDK5R1-bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Cyclin-dependent kinase 5 activator 1, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-22 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
4XCP
 
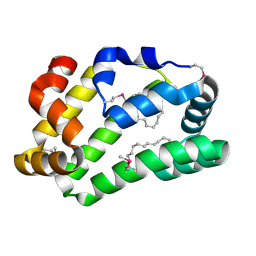 | | Fatty Acid and Retinol binding protein Na-FAR-1 from Necator americanus | | Descriptor: | Nematode fatty acid retinoid binding protein, PALMITIC ACID | | Authors: | Gabrielsen, M, Rey-Burusco, M.F, Ibanez-Shimabukuro, M, Griffiths, K, Kennedy, M.W, Corsico, B, Smith, B.O. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Diversity in the structures and ligand-binding sites of nematode fatty acid and retinol-binding proteins revealed by Na-FAR-1 from Necator americanus.
Biochem.J., 471, 2015
|
|
4UIJ
 
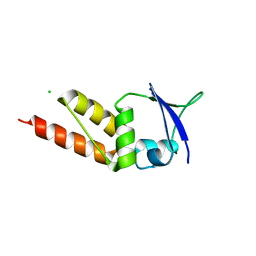 | | Crystal structure of the BTB domain of KCTD13 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN 1, CHLORIDE ION | | Authors: | Pinkas, D.M, Sanvitale, C.E, Sorell, F.J, Solcan, N, Goubin, S, Canning, P, Williams, E, Chaikuad, A, Dixon Clarke, S.E, Tallant, C, Fonseca, M, Chalk, R, Doutch, J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-03-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
8X7B
 
 | | ThT-bound E46K alpha-synuclein fibrils | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-23 | | Release date: | 2024-11-27 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6LEN
 
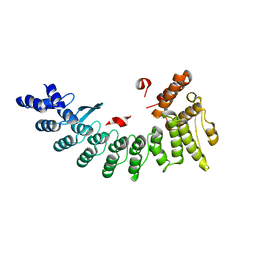 | | Structure of NS11 bound FEM1C | | Descriptor: | Protein fem-1 homolog C,NS11 peptide | | Authors: | Chen, x, Liao, S, Xu, C. | | Deposit date: | 2019-11-25 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
1IQC
 
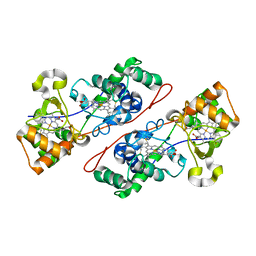 | | Crystal structure of Di-Heme Peroxidase from Nitrosomonas europaea | | Descriptor: | CALCIUM ION, GLYCEROL, HEME C, ... | | Authors: | Shimizu, H. | | Deposit date: | 2001-07-20 | | Release date: | 2002-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Nitrosomonas europaea cytochrome c peroxidase and the structural basis for ligand switching in bacterial di-heme peroxidases
Biochemistry, 40, 2001
|
|
5D5R
 
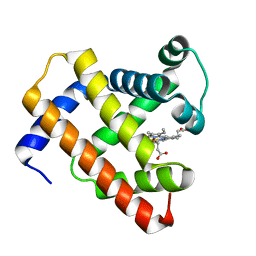 | | Horse-heart myoglobin - deoxy state | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Barends, T, Schlichting, I. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
1QI0
 
 | |
2J8T
 
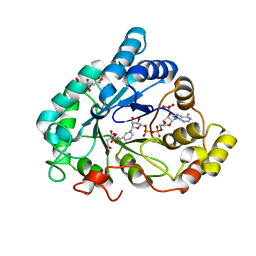 | | Human aldose reductase in complex with NADP and citrate at 0.82 angstrom | | Descriptor: | ALDO-KETO REDUCTASE FAMILY 1, MEMBER B1, CITRATE ANION, ... | | Authors: | Biadene, M, Hazemann, I, Cousido, A, Ginell, S, Sheldrick, G.M, Podjarny, A, Schneider, T.R. | | Deposit date: | 2006-10-27 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.82 Å) | | Cite: | The Atomic Resolution Structure of Human Aldose Reductase Reveals that Rearrangement of a Bound Ligand Allows the Opening of the Safety-Belt Loop.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3DGN
 
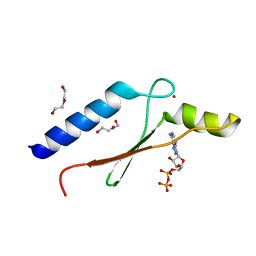 | | A non-biological ATP binding protein crystallized in the presence of 100 mM ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP Binding Protein-DX, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Simmons, C.R, Allen, J.P, Chaput, J.C. | | Deposit date: | 2008-06-13 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A synthetic protein selected for ligand binding affinity mediates ATP hydrolysis.
Acs Chem.Biol., 4, 2009
|
|
6TNO
 
 | | Crystal structure of the human Arc N-lobe bound to stargazin | | Descriptor: | Arc_C domain-containing protein, Chains: B,D,F | | Authors: | Hallin, E.I, Bramham, C.R, Kursula, P. | | Deposit date: | 2019-12-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural properties and peptide ligand binding of the capsid homology domains of human Arc.
Biochem Biophys Rep, 26, 2021
|
|
3DGL
 
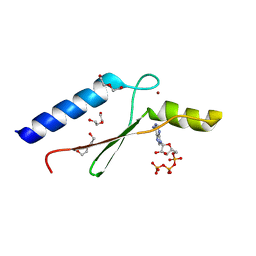 | | 1.8 A Crystal Structure of a Non-biological Protein with Bound ATP in a Novel Bent Conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP Binding Protein-DX, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Simmons, C.R, Allen, J.P, Chaput, J.C. | | Deposit date: | 2008-06-13 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A synthetic protein selected for ligand binding affinity mediates ATP hydrolysis.
Acs Chem.Biol., 4, 2009
|
|
6LBN
 
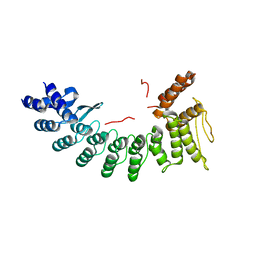 | | Structure of SIL1-bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Nucleotide exchange factor SIL1 | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LEY
 
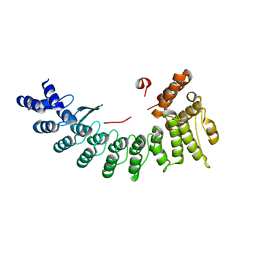 | | Structure of Sil1G bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Nucleotide exchange factor SIL1 | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6TNQ
 
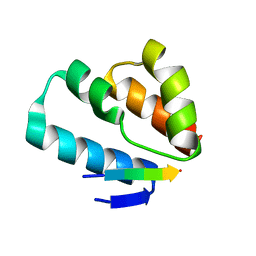 | | Crystal structure of the human Arc N-lobe bound to repeat 4 from GKAP | | Descriptor: | ACETYL GROUP, Activity-regulated cytoskeleton-associated protein, Chains: B,D,F | | Authors: | Hallin, E.I, Bramham, C.R, Kursula, P. | | Deposit date: | 2019-12-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural properties and peptide ligand binding of the capsid homology domains of human Arc.
Biochem Biophys Rep, 26, 2021
|
|
6FQG
 
 | | GluA2(flop) G724C ligand binding core dimer bound to L-Glutamate (Form A) at 2.34 Angstrom resolution | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.34139085 Å) | | Cite: | X-ray structure of GluA2 flop G724C ligand binding core dimer bound to glutamate at 2.32 Angstroms resolution
To Be Published
|
|
6LF0
 
 | | Structure of FEM1C | | Descriptor: | Protein fem-1 homolog C, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LA0
 
 | | Crystal structure of AoRut | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoside hydrolase family 5 | | Authors: | Koseki, T, Makabe, K. | | Deposit date: | 2019-11-11 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aspergillus oryzae Rutinosidase: Biochemical and Structural Investigation.
Appl.Environ.Microbiol., 87, 2021
|
|
7SHU
 
 | | IgE-Fc in complex with omalizumab variant C02 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant epsilon, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
6TN7
 
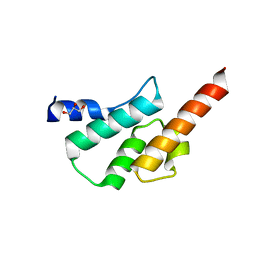 | | Crystal structure of the human Arc C-lobe | | Descriptor: | Activity-regulated cytoskeleton-associated protein, GLYCEROL | | Authors: | Hallin, E.I, Bramham, C.R, Kursula, P. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural properties and peptide ligand binding of the capsid homology domains of human Arc.
Biochem Biophys Rep, 26, 2021
|
|
1EUP
 
 | |
4A11
 
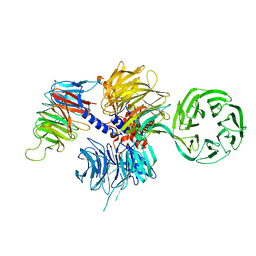 | | Structure of the hsDDB1-hsCSA complex | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, DNA EXCISION REPAIR PROTEIN ERCC-8 | | Authors: | Bohm, K, Scrima, A, Fischer, E.S, Gut, H, Thomae, N.H. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
3SV2
 
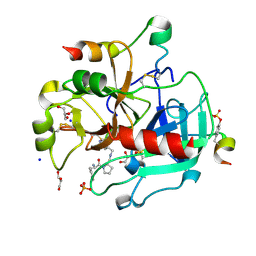 | | Human Thrombin In Complex With UBTHR105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-(pyridin-4-ylmethyl)-L-prolinamide, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-07-12 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
8JXT
 
 | | Histamine-bound H4R/Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2023-07-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of ligand recognition and design of antihistamines targeting histamine H 4 receptor.
Nat Commun, 15, 2024
|
|
8JXX
 
 | | Clobenpropit-bound H4R/Gi complex | | Descriptor: | 3-(1~{H}-imidazol-4-yl)propyl ~{N}'-[(4-chlorophenyl)methyl]carbamimidothioate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2023-07-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of ligand recognition and design of antihistamines targeting histamine H 4 receptor.
Nat Commun, 15, 2024
|
|
