7SAS
 
 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
7SAQ
 
 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
7SAR
 
 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
7QIQ
 
 | | CRYSTAL STRUCTURE OF THE P1 aminobutanoic acid (ABU) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
7QIR
 
 | | CRYSTAL STRUCTURE OF THE P1 monofluorethylglycine(MfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
7QIS
 
 | | CRYSTAL STRUCTURE OF THE P1 difluoroethylglycine (DfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
7QIT
 
 | | CRYSTAL STRUCTURE OF THE P1 trifluoroethylglycine (TfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
7S7Q
 
 | |
7S1I
 
 | | Wild-type Escherichia coli stalled ribosome with antibiotic radezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S1J
 
 | | Wild-type Escherichia coli ribosome with antibiotic radezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S7R
 
 | | Plasmodium falciparum protein Pf12 bound to nanobody G7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Merozoite surface protein P12, Nanobody G7, ... | | Authors: | Dietrich, M.H, Tham, W.H. | | Deposit date: | 2021-09-17 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of the Pf12 and Pf41 heterodimeric complex of Plasmodium falciparum 6-cysteine proteins.
FEMS Microbes, 3, 2022
|
|
7MQQ
 
 | |
7NRY
 
 | | Re-refinement of MAPKAP kinase-2/inhibitor complex 3fyj | | Descriptor: | (10R)-10-methyl-3-(6-methylpyridin-3-yl)-9,10,11,12-tetrahydro-8H-[1,4]diazepino[5',6':4,5]thieno[3,2-f]quinolin-8-one, CHLORIDE ION, MALONIC ACID, ... | | Authors: | Croll, T.I, Read, R.J. | | Deposit date: | 2021-03-04 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Adaptive Cartesian and torsional restraints for interactive model rebuilding.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7LHQ
 
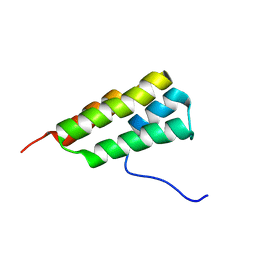 | | Solution structure of SARS-CoV-2 nonstructural protein 7 at pH 7.0 | | Descriptor: | Non-structural protein 7 | | Authors: | Lee, Y, Tonelli, M, Anderson, T.K, Kirchdoerfer, R.N, Henzler-Wildman, K, Lee, W. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-dependent polymorphism of the structure of SARS-CoV-2 nsp7
Biorxiv, 2021
|
|
7Q5G
 
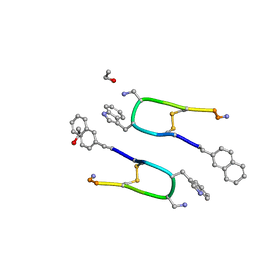 | | LAN-DAP5 DERIVATIVE OF LANREOTIDE: L-DIAMINO PROPIONIC ACID IN POSITION 5 IN PLACE OF L-LYSINE | | Descriptor: | ETHANOL, LAN-DAP5 DERIVATIVE OF LANREOTIDE | | Authors: | Bressanelli, S, Le Du, M.H, Gobeaux, F, Legrand, P, Paternostre, M. | | Deposit date: | 2021-11-03 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Atomic structure of Lanreotide nanotubes revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RY7
 
 | |
7NDT
 
 | | UL40:01 TCR in complex with HLA-E with a non-natural amino acid | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
7NDU
 
 | | Gag:02 TCR in complex with HLA-E featuring a non-natural amino acid | | Descriptor: | Beta-2-microglobulin, Gag6V(276-284 H4C), HLA class I histocompatibility antigen, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
7NDQ
 
 | | Gag:02 TCR in complex with HLA-E. | | Descriptor: | Beta-2-microglobulin, Gag6V, HLA class I histocompatibility antigen, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
7QBN
 
 | | Structure of cathepsin K in complex with the azadipeptide nitrile inhibitor Gu1303 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[aminomethyl(methyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7QBL
 
 | |
7QBM
 
 | | Structure of the activation intermediate of cathepsin K in complex with the 3-cyano-3-aza-beta-amino acid inhibitor Gu2602 | | Descriptor: | ACETATE ION, Cathepsin K, MAGNESIUM ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7QBO
 
 | | Structure of the activation intermediate of cathepsin K in complex with the azadipeptide nitrile inhibitor Gu1303 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[aminomethyl(methyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
6ZKY
 
 | | Crystal structure of InhA:01 TCR in complex with HLA-E (S147C) bound to InhA (53-61 H3C) | | Descriptor: | Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], HLA class I histocompatibility antigen, ... | | Authors: | Srikannathasan, V, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2020-06-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
6ZKW
 
 | | Crystal structure of InhA:01 TCR in complex with HLA-E bound to InhA (53-61) | | Descriptor: | Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], HLA class I histocompatibility antigen, ... | | Authors: | Srikannathasan, V, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2020-06-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
