5LEL
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_10_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | Descriptor: | DD_Off7_10_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-30 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5J3N
 
 | | C-terminal domain of EcoR124I HsdR subunit fused with the pH-sensitive GFP variant ratiometric pHluorin | | Descriptor: | Green fluorescent protein,HsdR | | Authors: | Grinkevich, P, Iermak, I, Luedtke, N, Mesters, J.R, Ettrich, R, Ludwig, J. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a novel domain of the motor subunit of the Type I restriction enzyme EcoR124 involved in complex assembly and DNA binding.
J. Biol. Chem., 293, 2018
|
|
5LEM
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_11_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | Descriptor: | DD_Off7_11_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
8C7I
 
 | |
7VCM
 
 | | crystal structure of GINKO1 | | Descriptor: | Green fluorescent protein,Potassium binding protein Kbp,Green fluorescent protein, POTASSIUM ION | | Authors: | Wen, Y, Campbell, R.E, Lemieux, M.J. | | Deposit date: | 2021-09-03 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A sensitive and specific genetically-encoded potassium ion biosensor for in vivo applications across the tree of life.
Plos Biol., 20, 2022
|
|
8DN2
 
 | |
8DN4
 
 | | Cryo-EM structure of human Glycine Receptor alpha-1 beta heteromer, glycine-bound state3(desensitized state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-1, Glycine receptor subunit beta,Green fluorescent protein,Glycine receptor beta, ... | | Authors: | Liu, X, Wang, W. | | Deposit date: | 2022-07-10 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric gating of a human hetero-pentameric glycine receptor.
Nat Commun, 14, 2023
|
|
8DN5
 
 | |
8DN3
 
 | |
6B0B
 
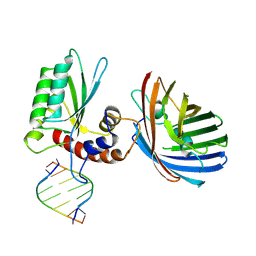 | | Crystal structure of human APOBEC3H | | Descriptor: | APOBEC3H, MCherry, RNA (5'-R(*UP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Shaban, N.M, Shi, K, Banerjee, S, Harris, R.S, Aihara, H. | | Deposit date: | 2017-09-14 | | Release date: | 2017-10-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.2800622 Å) | | Cite: | The Antiviral and Cancer Genomic DNA Deaminase APOBEC3H Is Regulated by an RNA-Mediated Dimerization Mechanism.
Mol. Cell, 69, 2018
|
|
6BBO
 
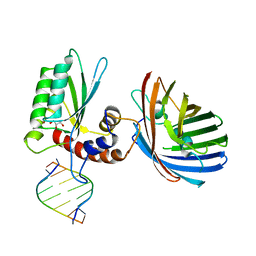 | | Crystal structure of human APOBEC3H/RNA complex | | Descriptor: | APOBEC3H, GLYCEROL, MCherry fluorescent protein, ... | | Authors: | Shaban, N.M, Shi, K, Banerjee, S, Harris, R.S, Aihara, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.428 Å) | | Cite: | The Antiviral and Cancer Genomic DNA Deaminase APOBEC3H Is Regulated by an RNA-Mediated Dimerization Mechanism.
Mol. Cell, 69, 2018
|
|
7Y96
 
 | |
3OSQ
 
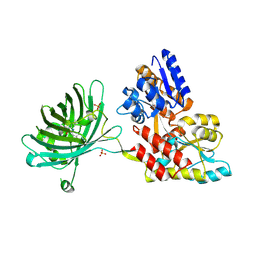 | | Maltose-bound maltose sensor engineered by insertion of circularly permuted green fluorescent protein into E. coli maltose binding protein at position 175 | | Descriptor: | Maltose-binding periplasmic protein,Green fluorescent protein, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Echevarria, I.M, Marvin, J.S, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2010-09-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A genetically encoded, high-signal-to-noise maltose sensor.
Proteins, 79, 2011
|
|
3O78
 
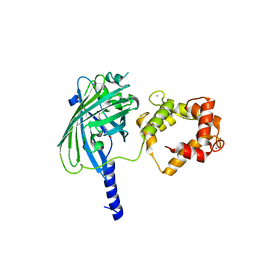 | | The structure of Ca2+ Sensor (Case-12) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, smooth muscle,Green fluorescent protein,Green fluorescent protein,Calmodulin-1 | | Authors: | Leder, L, Stark, W, Freuler, F, Marsh, M, Meyerhofer, M, Stettler, T, Mayr, L.M, Britanova, O.V, Strukova, L.A, Chudakov, D.M. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of Ca2+ sensor Case16 reveals the mechanism of reaction to low Ca2+ concentrations
Sensors (Basel), 10, 2010
|
|
3EVU
 
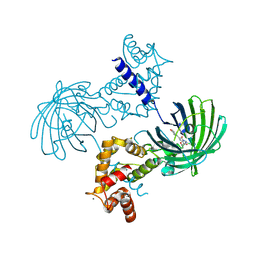 | | Crystal structure of Calcium bound dimeric GCAMP2 | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Wang, Q, Shui, B, Kotlikoff, M.I, Sondermann, H. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Calcium Sensing by GCaMP2.
Structure, 16, 2008
|
|
3EVV
 
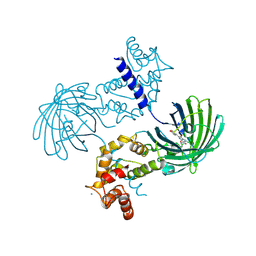 | | Crystal Structure of Calcium bound dimeric GCAMP2 (#2) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Wang, Q, Shui, B, Kotlikoff, M.I, Sondermann, H. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Calcium Sensing by GCaMP2.
Structure, 16, 2008
|
|
3OSR
 
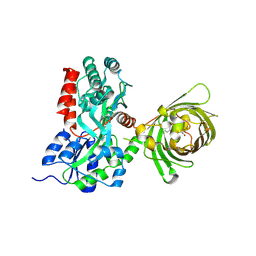 | | Maltose-bound maltose sensor engineered by insertion of circularly permuted green fluorescent protein into E. coli maltose binding protein at position 311 | | Descriptor: | Maltose-binding periplasmic protein,Green fluorescent protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Echevarria, I.M, Marvin, J.S, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2010-09-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A genetically encoded, high-signal-to-noise maltose sensor.
Proteins, 79, 2011
|
|
3O77
 
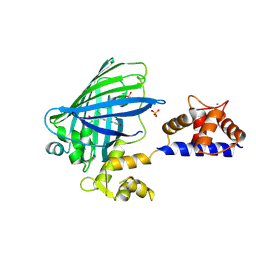 | | The structure of Ca2+ Sensor (Case-16) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Myosin light chain kinase, ... | | Authors: | Leder, L, Stark, W, Freuler, F, Marsh, M, Meyerhofer, M, Stettler, T, Mayr, L.M, Britanova, O.V, Strukova, L.A, Chudakov, D.M. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of Ca2+ sensor Case16 reveals the mechanism of reaction to low Ca2+ concentrations
Sensors (Basel), 10, 2010
|
|
7CXR
 
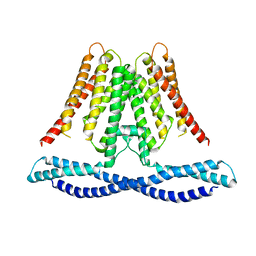 | | Cryo-EM structure of human TMEM120A/TACAN | | Descriptor: | MCherry fluorescent protein,Ion channel TACAN | | Authors: | Yan, Z, Wu, J, Ke, M. | | Deposit date: | 2020-09-02 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human TMEM120A and TMEM120B.
Cell Discov, 7, 2021
|
|
7EDJ
 
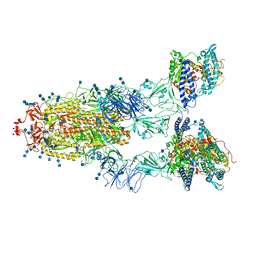 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-01 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7BWN
 
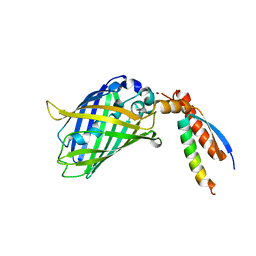 | | Crystal Structure of a Designed Protein Heterocatenane | | Descriptor: | Cellular tumor antigen p53, Chimera of Green fluorescent protein and p53dim | | Authors: | Liu, Y.J, Duan, Z.L, Fang, J, Zhang, F, Xiao, J.Y, Zhang, W.B. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Cellular Synthesis and X-ray Crystal Structure of a Designed Protein Heterocatenane.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3AI4
 
 | | Crystal structure of yeast enhanced green fluorescent protein - mouse polymerase iota ubiquitin binding motif fusion protein | | Descriptor: | SULFATE ION, yeast enhanced green fluorescent protein,DNA polymerase iota | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6GEL
 
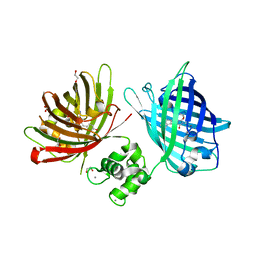 | | The structure of TWITCH-2B | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-26 | | Release date: | 2019-08-21 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
4XBI
 
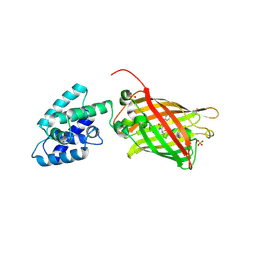 | | Structure Of A Malarial Protein Involved in Proteostasis | | Descriptor: | ClpB protein, putative,Green fluorescent protein, SULFATE ION | | Authors: | Egea, P.F, Ah Young, A.P, Cascio, D. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Structural mapping of the ClpB ATPases of Plasmodium falciparum: Targeting protein folding and secretion for antimalarial drug design.
Protein Sci., 24, 2015
|
|
4Z4K
 
 | |
