1MMW
 
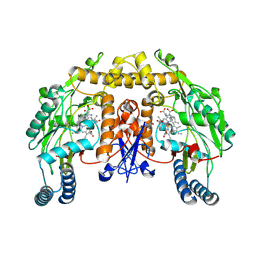 | | Rat neuronal NOS heme domain with vinyl-L-NIO bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N5-(1-IMINO-3-BUTENYL)-L-ORNITHINE, ... | | Authors: | Bretscher, L.E, Li, H, Poulos, T.L, Griffith, O.W. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization and kinetics of nitric-oxide synthase inhibition by novel N5-(iminoalkyl)- and N5-(iminoalkenyl)-ornithines
J.Biol.Chem., 278, 2003
|
|
6RFD
 
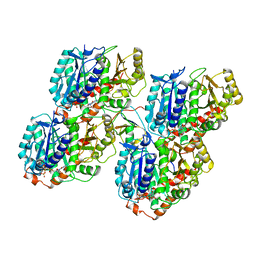 | |
6BEC
 
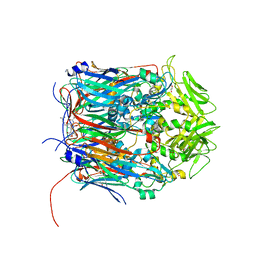 | | Crystal structure of VACV D13 in complex with Rifabutin | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, RIFABUTIN, ... | | Authors: | Garriga, D, Accurso, C, Coulibaly, F. | | Deposit date: | 2017-10-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural basis for the inhibition of poxvirus assembly by the antibiotic rifampicin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5A5B
 
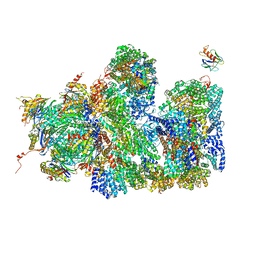 | | Structure of the 26S proteasome-Ubp6 complex | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Aufderheide, A, Beck, F, Stengel, F, Hartwig, M, Schweitzer, A, Pfeifer, G, Goldberg, A.L, Sakata, E, Baumeister, W, Foerster, F. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-22 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structural Characterization of the Interaction of Ubp6 with the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1NWK
 
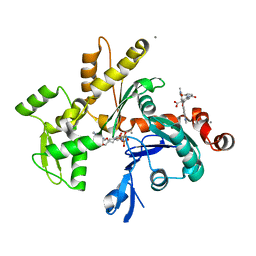 | | CRYSTAL STRUCTURE OF MONOMERIC ACTIN IN THE ATP STATE | | Descriptor: | Actin, alpha skeletal muscle, CALCIUM ION, ... | | Authors: | Graceffa, P, Dominguez, R. | | Deposit date: | 2003-02-06 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics.
J.Biol.Chem., 278, 2003
|
|
3GZY
 
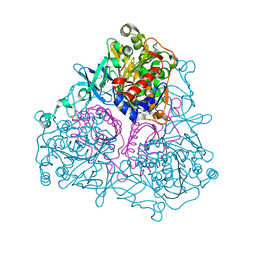 | | Crystal Structure of the Biphenyl Dioxygenase from Comamonas testosteroni Sp. Strain B-356 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Biphenyl dioxygenase subunit alpha, Biphenyl dioxygenase subunit beta, ... | | Authors: | Kumar, P, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2009-04-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Characterization of Pandoraea pnomenusa B-356 Biphenyl Dioxygenase Reveals Features of Potent Polychlorinated Biphenyl-Degrading Enzymes
Plos One, 8, 2013
|
|
1M8X
 
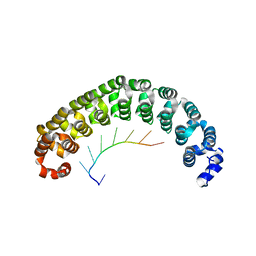 | | CRYSTAL STRUCTURE OF THE PUMILIO-HOMOLOGY DOMAIN FROM HUMAN PUMILIO1 IN COMPLEX WITH NRE1-14 RNA | | Descriptor: | 5'-R(P*UP*GP*UP*AP*UP*AP*U)-3', 5'-R(P*UP*UP*GP*UP*AP*UP*AP*U)-3', Pumilio 1 | | Authors: | Wang, X, McLachlan, J, Zamore, P.D, Hall, T.M.T. | | Deposit date: | 2002-07-26 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MODULAR RECOGNITION OF RNA BY A HUMAN PUMILIO-HOMOLOGY DOMAIN
CELL(CAMBRIDGE,MASS.), 110, 2002
|
|
6HWV
 
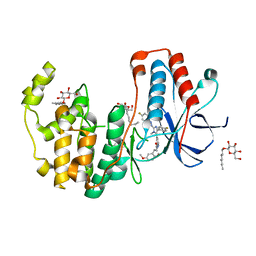 | | Crystal structure of p38alpha in complex with a photoswitchable 2-Azoimidazol-based Inhibitor (compound 3) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[5-(4-fluorophenyl)-2-[(~{E})-(4-fluorophenyl)diazenyl]-3-methyl-imidazol-4-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Mueller, M.P, Rauh, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
1MB6
 
 | |
3MVR
 
 | |
4YHH
 
 | | Crystal structure of the 30S ribosomal subunit from Thermus thermophilus in complex with tigecycline | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Kaminishi, T, Ochoa-Lizarralde, B, Dhimole, N, Zhou, S, Lopez-Alonso, J.P, Connell, S.R, Fucini, P. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.417 Å) | | Cite: | Structural characterization of an alternative mode of tigecycline binding to the bacterial ribosome.
Antimicrob.Agents Chemother., 59, 2015
|
|
1P3H
 
 | | Crystal Structure of the Mycobacterium tuberculosis chaperonin 10 tetradecamer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 10 kDa chaperonin, CALCIUM ION | | Authors: | Roberts, M.M, Coker, A.R, Fossati, G, Mascagni, P, Coates, A.R.M, Wood, S.P, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis chaperonin 10 heptamers self-associate through their biologically active loops
J.BACTERIOL., 185, 2003
|
|
5AEU
 
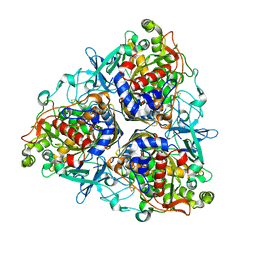 | | Crystal structure of II9 variant of Biphenyl dioxygenase from Burkholderia xenovorans LB400 | | Descriptor: | BIPHENYL DIOXYGENASE SUBUNIT ALPHA, BIPHENYL DIOXYGENASE SUBUNIT BETA, FE (II) ION, ... | | Authors: | Dhindwal, S, Gomez-Gil, L, Sylvestre, M, Eltis, L.D, Bolin, J.T, Kumar, P. | | Deposit date: | 2015-01-10 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Basis of the Enhanced Pollutant-Degrading Capabilities of an Engineered Biphenyl Dioxygenase
J.Bacteriol., 198, 2016
|
|
3H1I
 
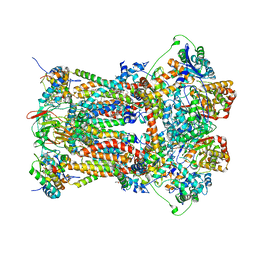 | | Stigmatellin and antimycin bound cytochrome bc1 complex from chicken | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-METHYL-BUTYRIC ACID 3-(3-FORMYLAMINO-2-HYDROXY-BENZOYLAMINO)-8-HEPTYL-2,6-DIMETHYL-4,9-DIOXO-[1,5]DIOXONAN-7-YL ESTER, CARDIOLIPIN, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
6RD7
 
 | | CryoEM structure of Polytomella F-ATP synthase, c-ring position 1, focussed refinement of Fo and peripheral stalk | | Descriptor: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ASA-9: Polytomella F-ATP synthase associated subunit 9, ATP synthase associated protein ASA1, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDU
 
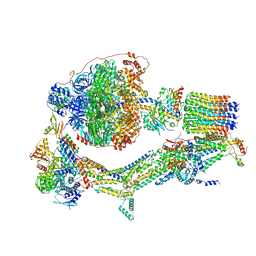 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1E, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6REC
 
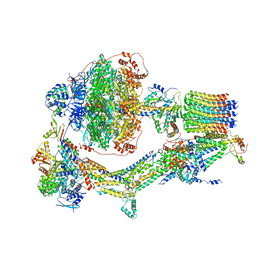 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3A, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
3H1J
 
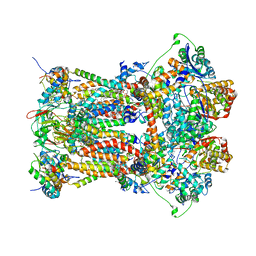 | | Stigmatellin-bound cytochrome bc1 complex from chicken | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
1OHH
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE complexed with the inhibitor protein IF1 | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Cabezon, E, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of Bovine F1-ATPase in Complex with its Regulatory Protein If1
Nat.Struct.Biol., 10, 2003
|
|
6SO2
 
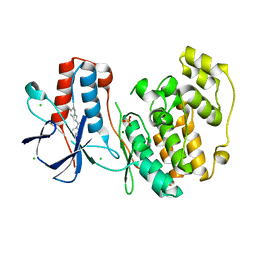 | | Fragment N13460a in complex with MAP kinase p38-alpha | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mitogen-activated protein kinase 14, ... | | Authors: | Nichols, C.E, De Nicola, G.F. | | Deposit date: | 2019-08-28 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
6SPL
 
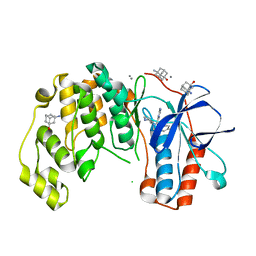 | | Fragment KCL615 in complex with MAP kinase p38-alpha | | Descriptor: | (5~{S},7~{R})-3-azanyladamantan-1-ol, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, CALCIUM ION, ... | | Authors: | Nichols, C.E, De Nicola, G.F. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
4X6Y
 
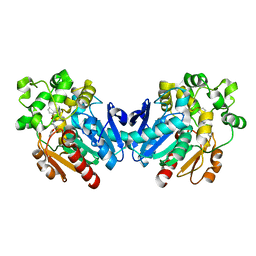 | |
3IBD
 
 | | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Gay, S.C, Sun, L, Talakad, J.C, Shah, M.B, Stout, D.C, Halpert, J.R. | | Deposit date: | 2009-07-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-A resolution.
Mol.Pharmacol., 77, 2010
|
|
5ARH
 
 | | Bovine mitochondrial ATP synthase state 2a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
5ARA
 
 | | Bovine mitochondrial ATP synthase state 1a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
