4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | Descriptor: | Eosinophil cationic protein, SULFATE ION | | Authors: | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
4RCM
 
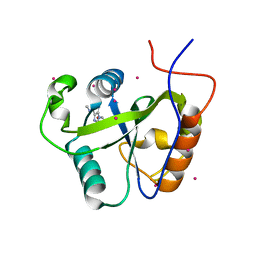 | | Crystal structure of the Pho92 YTH domain in complex with m6A | | Descriptor: | Methylated RNA-binding protein 1, RNA (5'-R(*UP*G)-D(*(6MZ)P*CP*U)-3'), UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Xu, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
3SFU
 
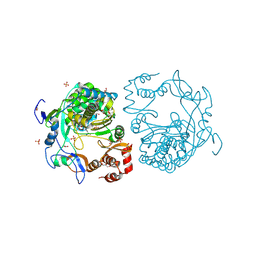 | | crystal structure of murine norovirus RNA dependent RNA polymerase in complex with ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, K.H, Alam, I. | | Deposit date: | 2011-06-14 | | Release date: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase in complex with 2-thiouridine or ribavirin.
Virology, 426, 2012
|
|
8QMA
 
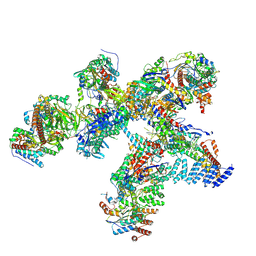 | | Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | do Prado, P.F.V, Ahrens, F.M, Pfannschmidt, T, Hillen, H.S. | | Deposit date: | 2023-09-21 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the multi-subunit chloroplast RNA polymerase.
Mol.Cell, 84, 2024
|
|
6HAY
 
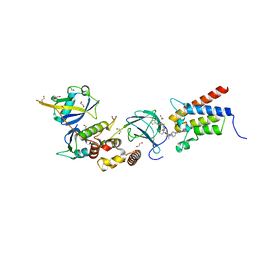 | | Crystal structure of PROTAC 1 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[2-[2-[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]ethoxy]ethoxy]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
2CPN
 
 | | Solution structure of the second dsRBD of TAR RNA-binding protein 2 | | Descriptor: | TAR RNA-binding protein 2 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second dsRBD of TAR RNA-binding protein 2
To be Published
|
|
6W5C
 
 | | Cryo-EM structure of Cas12i(E894A)-crRNA-dsDNA complex | | Descriptor: | Cas12i, NTS, Substrate, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanisms for target recognition and cleavage by the Cas12i RNA-guided endonuclease.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2AO5
 
 | | Crystal structure of an RNA duplex r(GGCGBrUGCGCU)2 with terminal and internal tandem G-U base pairs | | Descriptor: | 5'-R(*GP*GP*CP*GP*(5BU)P*GP*CP*GP*CP*U)-3', MAGNESIUM ION | | Authors: | Utsunomiya, R, Suto, K, Balasundaresan, D, Fukamizu, A, Kumar, P.K, Mizuno, H. | | Deposit date: | 2005-08-12 | | Release date: | 2006-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an RNA duplex r(GGCGBrUGCGCU)2 with terminal and internal tandem G.U base pairs.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1DM9
 
 | | HEAT SHOCK PROTEIN 15 KD | | Descriptor: | HYPOTHETICAL 15.5 KD PROTEIN IN MRCA-PCKA INTERGENIC REGION, SULFATE ION | | Authors: | Staker, B.L, Korber, P, Bardwell, J.C.A, Saper, M.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Hsp15 reveals a novel RNA-binding motif.
EMBO J., 19, 2000
|
|
6HAX
 
 | | Crystal structure of PROTAC 2 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[4-[[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]methyl]phenyl]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
6CMN
 
 | | Co-Crystal Structure of HIV-1 TAR Bound to Lab-Evolved RRM TBP6.7 | | Descriptor: | TAR-Binding Protein 6.7, Trans-Activation Response RNA Element | | Authors: | Belashov, I.A, Wedekind, J.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structure of HIV TAR in complex with a Lab-Evolved RRM provides insight into duplex RNA recognition and synthesis of a constrained peptide that impairs transcription.
Nucleic Acids Res., 46, 2018
|
|
1ARO
 
 | | T7 RNA POLYMERASE COMPLEXED WITH T7 LYSOZYME | | Descriptor: | MERCURY (II) ION, T7 LYSOZYME, T7 RNA POLYMERASE | | Authors: | Steitz, T, Jeruzalmi, D. | | Deposit date: | 1997-08-08 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme.
EMBO J., 17, 1998
|
|
4J39
 
 | |
4NRT
 
 | | Human Norovirus polymerase bound to Compound 6 (suramin derivative) | | Descriptor: | 4-({4-methyl-3-[(3-nitrobenzoyl)amino]benzoyl}amino)naphthalene-1,5-disulfonic acid, hNV-RdRp | | Authors: | Croci, R, Pezzullo, M, Tarantino, D, Mastrangelo, E, Milani, M, Bolognesi, M. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds.
Plos One, 9, 2014
|
|
4JNX
 
 | |
2F5W
 
 | | Cross-linked barnase soaked in 3 M thiourea | | Descriptor: | Ribonuclease, THIOUREA, ZINC ION | | Authors: | Prange, T, Salem, M. | | Deposit date: | 2005-11-28 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | On the edge of the denaturation process: Application of X-ray diffraction to barnase and lysozyme cross-linked crystals with denaturants in molar concentrations.
Biochim.Biophys.Acta, 1764, 2006
|
|
4CKB
 
 | | Vaccinia virus capping enzyme complexed with GTP and SAH | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, ... | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
4JVH
 
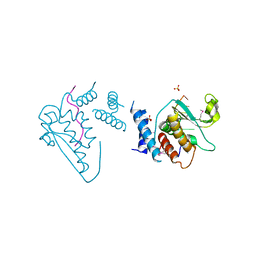 | | Structure of the star domain of quaking protein in complex with RNA | | Descriptor: | Protein quaking, RNA (5'-R(*UP*UP*CP*AP*CP*UP*AP*AP*CP*AP*A)-3'), SULFATE ION | | Authors: | Teplova, M, Hafner, M, Teplov, D, Essig, K, Tuschl, T, Patel, D.J. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites.
Genes Dev., 27, 2013
|
|
2F5M
 
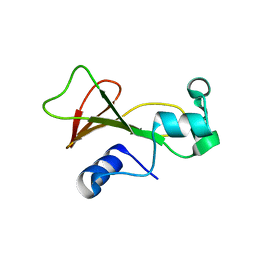 | | Cross-linked barnase soaked in bromo-ethanol | | Descriptor: | 2-BROMOETHANOL, Ribonuclease | | Authors: | Prange, T, Salem, M. | | Deposit date: | 2005-11-26 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | On the edge of the denaturation process: Application of X-ray diffraction to barnase and lysozyme cross-linked crystals with denaturants in molar concentrations.
Biochim.Biophys.Acta, 1764, 2006
|
|
4JVY
 
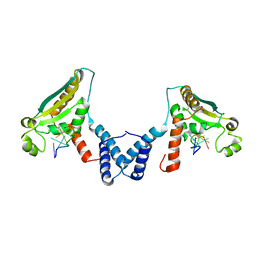 | | Structure of the STAR (signal transduction and activation of RNA) domain of GLD-1 bound to RNA | | Descriptor: | Female germline-specific tumor suppressor gld-1, RNA (5'-R(P*CP*UP*AP*AP*CP*AP*A)-3') | | Authors: | Teplova, M, Hafner, M, Teplov, D, Essig, K, Tuschl, T, Patel, D.J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-08 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites.
Genes Dev., 27, 2013
|
|
3CRR
 
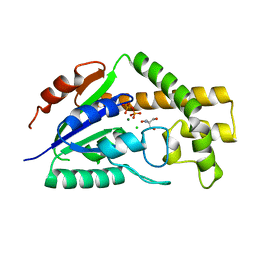 | | Structure of tRNA Dimethylallyltransferase: RNA Modification through a Channel | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIPHOSPHATE, ... | | Authors: | Huang, R.H, Xie, W, Zhou, C. | | Deposit date: | 2008-04-07 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of tRNA Dimethylallyltransferase: RNA Modification
through a Channel
J.Mol.Biol., 367, 2007
|
|
1X1P
 
 | | Crystal structure of Tk-RNase HII(1-197)-A(28-42) | | Descriptor: | Ribonuclease HII | | Authors: | Takano, K, Endo, S, Mukaiyama, A, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2005-04-11 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of amyloid beta fragments in aqueous environments
Febs J., 273, 2006
|
|
5EOV
 
 | |
6LU0
 
 | | Crystal structure of Cas12i2 ternary complex with 12 nt spacer | | Descriptor: | Cas12i2, DNA (5'-D(*GP*CP*CP*GP*CP*TP*TP*TP*CP*TP*T)-3'), DNA (5'-D(*GP*CP*TP*TP*GP*CP*TP*CP*TP*GP*TP*TP*GP*AP*AP*AP*GP*CP*GP*GP*C)-3'), ... | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-24 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
5WTI
 
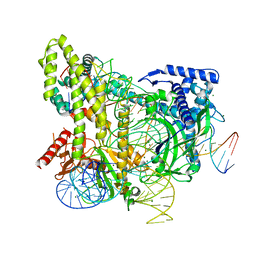 | | Crystal structure of the CRISPR-associated protein in complex with crRNA and DNA | | Descriptor: | CRISPR-associated protein, DNA (28-MER), DNA (5'-D(P*GP*TP*GP*TP*GP*GP*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Wu, D, Guan, X, Zhu, Y, Huang, Z. | | Deposit date: | 2016-12-13 | | Release date: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Structural basis of stringent PAM recognition by CRISPR-C2c1 in complex with sgRNA
Cell Res., 27, 2017
|
|
