2QKW
 
 | | Structural basis for activation of plant immunity by bacterial effector protein AvrPto | | Descriptor: | Avirulence protein, Protein kinase | | Authors: | Xing, W.M, Zou, Y, Liu, Q, Hao, Q, Zhou, J.M, Chai, J.J. | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis for activation of plant immunity by bacterial effector protein AvrPto
Nature, 449, 2007
|
|
1H9O
 
 | | PHOSPHATIDYLINOSITOL 3-KINASE, P85-ALPHA SUBUNIT: C-TERMINAL SH2 DOMAIN COMPLEXED WITH A TYR751 PHOSPHOPEPTIDE FROM THE PDGF RECEPTOR, CRYSTAL STRUCTURE AT 1.79 A | | Descriptor: | BETA-PLATELET-DERIVED GROWTH FACTOR RECEPTOR, PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Pauptit, R.A, Rowsell, S, Breeze, A.L, Murshudov, G.N, Dennis, C.A, Derbyshire, D.J, Weston, S.A. | | Deposit date: | 2001-03-14 | | Release date: | 2001-03-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | NMR Trial Models: Experiences with the Colicin Immunity Protein Im7 and the P85Alpha C-Terminal Sh2-Peptide Complex
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2QCB
 
 | | T7-tagged full-length streptavidin complexed with ruthenium ligand | | Descriptor: | N-(4-{[(2-AMINOETHYL)AMINO]SULFONYL}PHENYL)-5-[(3AS,4S,6AR)-2-OXOHEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL]PENTANAMIDE-(1,2,3,4,5,6-ETA)-BENZENE-CHLORO-RUTHENIUM(III), Streptavidin | | Authors: | Le Trong, I, Creus, M, Pordea, A, Ward, T.R, Stenkamp, R.E. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and designed evolution of an artificial transfer hydrogenase
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
4N1P
 
 | |
2QY1
 
 | | pectate lyase A31G/R236F from Xanthomonas campestris | | Descriptor: | PHOSPHATE ION, Pectate lyase II | | Authors: | Garron, M.L, Shaya, D. | | Deposit date: | 2007-08-13 | | Release date: | 2008-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Improvement of the thermostability and activity of a pectate lyase by single amino acid substitutions, using a strategy based on melting-temperature-guided sequence alignment.
Appl.Environ.Microbiol., 74, 2008
|
|
2QZL
 
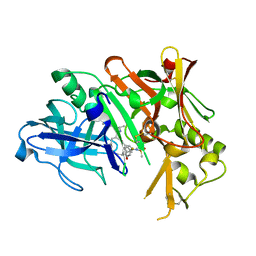 | | Crystal Structure of human Beta Secretase complexed with IXS | | Descriptor: | Beta-secretase 1, N-[(1S)-1-benzyl-2-{[(1S)-2-(isobutylamino)-1-methyl-2-oxoethyl]amino}ethyl]-N'-[(1R)-1-(4-fluorophenyl)ethyl]-5-[methyl(methylsulfonyl)amino]isophthalamide | | Authors: | Munshi, S. | | Deposit date: | 2007-08-16 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BACE-1 inhibition by a series of psi[CH2NH] reduced amide isosteres
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4N1E
 
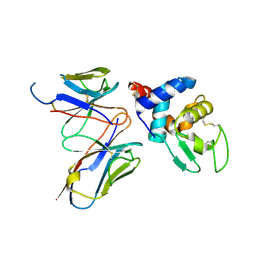 | | Structural evidence for antigen receptor evolution | | Descriptor: | Lysozyme C, immunoglobulin variable light chain domain | | Authors: | Langley, D.B, Rouet, R, Stock, D, Christ, D. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural reconstruction of protein ancestry.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4N1R
 
 | |
9CEA
 
 | |
9CDW
 
 | | Crystal structure of HP1alpha chromoshadow domain in complex with KAP1 peptide | | Descriptor: | Chromobox protein homolog 5, TRIETHYLENE GLYCOL, Transcription intermediary factor 1-beta peptide | | Authors: | Selvam, K, Singh, R.K, Gaurav, N, Kutateladze, T.G. | | Deposit date: | 2024-06-25 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The HP1 box of KAP1 organizes HP1 alpha for silencing of endogenous retroviral elements in embryonic stem cells.
Nat Commun, 16, 2025
|
|
4N9A
 
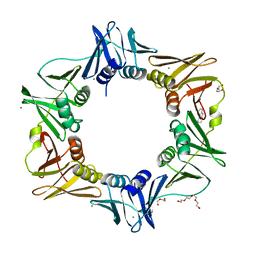 | | E. coli sliding clamp in complex with (R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid | | Descriptor: | (1R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4P8E
 
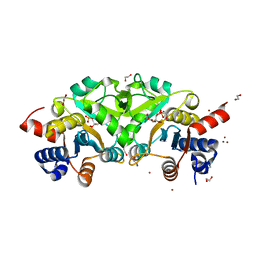 | | Structure of ribB complexed with substrate (Ru5P) and metal ions | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
1H0M
 
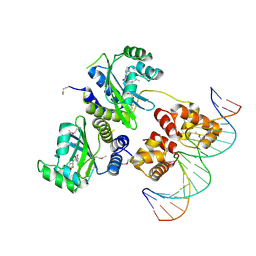 | | Three-dimensional structure of the quorum sensing protein TraR bound to its autoinducer and to its target DNA | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, 5'-D(*AP*TP*GP*TP*GP*CP*AP*GP*AP*TP *CP*TP*GP*CP*AP*CP*AP*T)-3', Transcriptional activator protein TraR | | Authors: | Vannini, A, Volpari, C, Gargioli, C, Muraglia, E, Cortese, R, De Francesco, R, Neddermann, P, Di Marco, S. | | Deposit date: | 2002-06-25 | | Release date: | 2002-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Quorum Sensing Protein Trar Bound to its Autoinducer and Target DNA
Embo J., 21, 2002
|
|
4N30
 
 | |
4R6F
 
 | |
4REF
 
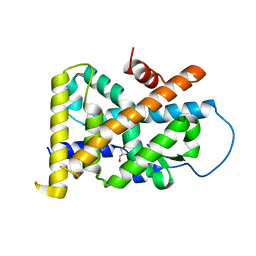 | | Crystal Structure of TR3 LBD_L449W in complex with Molecule 2 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)hexan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
2Q1C
 
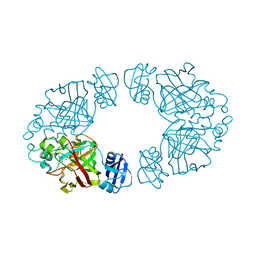 | | 2-keto-3-deoxy-D-arabinonate dehydratase complexed with calcium and 2-oxobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, 2-keto-3-deoxy-D-arabinonate dehydratase, CALCIUM ION | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
8V64
 
 | |
4NLK
 
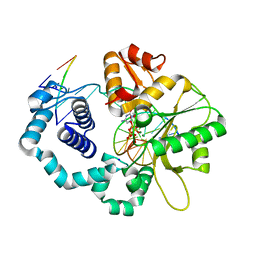 | | Structure of human DNA polymerase beta complexed with 8BrG in the template base-paired with incoming non-hydrolyzable CTP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, 5'-D(*CP*CP*GP*AP*CP*(BGM)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3', ... | | Authors: | Koag, M.-C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
6V23
 
 | |
2Q19
 
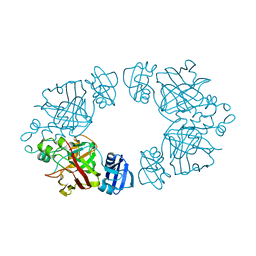 | | 2-keto-3-deoxy-D-arabinonate dehydratase apo form | | Descriptor: | 2-keto-3-deoxy-D-arabinonate dehydratase | | Authors: | Barends, T, Brouns, S, Worm, P, Akerboom, J, Turnbull, A, Salmon, L. | | Deposit date: | 2007-05-24 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-D-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily.
J.Mol.Biol., 379, 2008
|
|
8V60
 
 | | F315A mutant of human Slo1 in presence of EDTA - resting VSD | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL, Calcium-activated potassium channel subunit alpha-1, ... | | Authors: | Pal, K, Kallure, G, Chowdhury, S. | | Deposit date: | 2023-12-01 | | Release date: | 2024-12-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | F315A mutant of human Slo1 in presence of EDTA - resting VSD
To Be Published
|
|
6N2R
 
 | |
8VAV
 
 | | Human Slo1 - human LRRC26 in presence of EDTA - GR masked | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Pal, K, Kallure, G.S, Chowdhury, S. | | Deposit date: | 2023-12-11 | | Release date: | 2024-12-25 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Human Slo1 - human LRRC26 in presence of EDTA - GR masked
To Be Published
|
|
8VAZ
 
 | |
