6NEU
 
 | | FAD-dependent monooxygenase TropB from T. stipitatus R206Q variant | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6AGL
 
 | |
6O2N
 
 | |
6AGM
 
 | |
6O2M
 
 | |
6NEV
 
 | | FAD-dependent monooxygenase TropB from T. stipitatus Y239F Variant | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
1PNB
 
 | | STRUCTURE OF NAPIN BNIB, NMR, 10 STRUCTURES | | Descriptor: | NAPIN BNIB | | Authors: | Rico, M, Bruix, M, Gonzalez, C, Monsalve, R, Rodriguez, R. | | Deposit date: | 1996-09-17 | | Release date: | 1997-09-17 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | 1H NMR assignment and global fold of napin BnIb, a representative 2S albumin seed protein.
Biochemistry, 35, 1996
|
|
6O2O
 
 | |
7O83
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(7S)-11-chloro-12-(5-methyl-1H-indazol-4-yl)-9-oxa-2,5,15,17-tetrazatetracyclo[8.7.1.02,7.014,18]octadeca-1(17),10,12,14(18),15-pentaen-5-yl]prop-2-en-1-one, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2021-04-14 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
1KSI
 
 | | CRYSTAL STRUCTURE OF A EUKARYOTIC (PEA SEEDLING) COPPER-CONTAINING AMINE OXIDASE AT 2.2A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Wilce, M.C.J, Kumar, V, Freeman, H.C, Guss, J.M. | | Deposit date: | 1996-07-20 | | Release date: | 1997-12-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a eukaryotic (pea seedling) copper-containing amine oxidase at 2.2 A resolution.
Structure, 4, 1996
|
|
6OKU
 
 | |
6OKS
 
 | |
6OEF
 
 | | PolyAla Model of the O-layer from the Type 4 Secretion System of H. pylori | | Descriptor: | PolyAla Model of OMCC O-Layer | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6OKT
 
 | |
8D6J
 
 | |
8D71
 
 | | Human Ago2 bound to miR122(21nt) | | Descriptor: | MAGNESIUM ION, Protein argonaute-2, miR-122-21nt | | Authors: | Xiao, Y, MacRae, I. | | Deposit date: | 2022-06-07 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A tiny loop in the Argonaute PIWI domain tunes small RNA seed strength.
Embo Rep., 24, 2023
|
|
6O43
 
 | | Crystal structure of a lysin protein from Staphylococcus phage P68 | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Orf11 | | Authors: | Truong, J.Q. | | Deposit date: | 2019-02-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08222985 Å) | | Cite: | Combining random microseed matrix screening and the magic triangle for the efficient structure solution of a potential lysin from bacteriophage P68.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OEE
 
 | | Structure of CagT from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagT | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6OKR
 
 | |
5JNC
 
 | | Crystal structure for the complex of human carbonic anhydrase IV and 4-aminomethylbenzene sulfonamide | | Descriptor: | 4-(aminomethyl)benzene-1-sulfonamide, ACETATE ION, Carbonic anhydrase 4, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
5JNA
 
 | | Crystal structure for the complex of human carbonic anhydrase IV and topiramate | | Descriptor: | ACETATE ION, Carbonic anhydrase 4, GLYCEROL, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
8DU7
 
 | | Room-temperature serial synchrotron crystallography (SSX) structure of apo PTP1B | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Sharma, S, Keedy, D.A. | | Deposit date: | 2022-07-27 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room-temperature serial synchrotron crystallography of the human phosphatase PTP1B.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
6BNT
 
 | |
6PBB
 
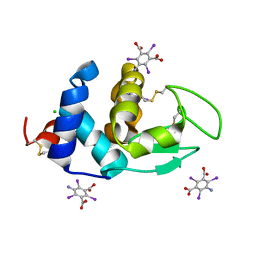 | | Crystal structure of Hen Egg White Lysozyme in complex with I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, Lysozyme | | Authors: | Truong, J.Q. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89151084 Å) | | Cite: | Combining random microseed matrix screening and the magic triangle for the efficient structure solution of a potential lysin from bacteriophage P68.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1M8Q
 
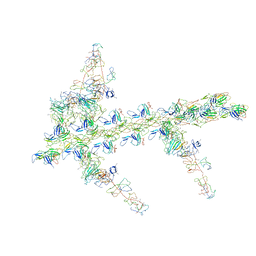 | | Molecular Models of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle | | Descriptor: | Skeletal muscle Actin, Skeletal muscle Myosin II, Skeletal muscle Myosin II Essential Light Chain, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-07-25 | | Release date: | 2002-09-10 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular modeling of averaged rigor crossbridges from tomograms of insect flight muscle.
J.Struct.Biol., 138, 2002
|
|
