4I1F
 
 | | Structure of Parkin-S223P E3 ligase | | Descriptor: | BARIUM ION, E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Lougheed, J.C, Brecht, E, Yao, N.H. | | Deposit date: | 2012-11-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases.
Nat Commun, 4, 2013
|
|
5WWX
 
 | | Crystal structure of the KH2 domain of human RNA-binding E3 ubiquitin-protein ligase MEX-3C complex with RNA | | Descriptor: | NICKEL (II) ION, RNA (5'-R(P*AP*GP*AP*GP*U)-3'), RNA-binding E3 ubiquitin-protein ligase MEX3C | | Authors: | Yang, L, Wang, C, Li, F, Gong, Q. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity
J. Biol. Chem., 292, 2017
|
|
5WWW
 
 | | Crystal structure of the KH1 domain of human RNA-binding E3 ubiquitin-protein ligase MEX-3C complex with RNA | | Descriptor: | RNA (5'-R(*GP*UP*UP*UP*AP*G)-3'), RNA-binding E3 ubiquitin-protein ligase MEX3C | | Authors: | Yang, L, Wang, C, Li, F, Gong, Q. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity
J. Biol. Chem., 292, 2017
|
|
5WWZ
 
 | | Crystal structure of the KH2 domain of human RNA-binding E3 ubiquitin-protein ligase MEX-3C | | Descriptor: | RNA-binding E3 ubiquitin-protein ligase MEX3C, SULFATE ION | | Authors: | Yang, L, Wang, C, Li, F, Gong, Q. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity
J. Biol. Chem., 292, 2017
|
|
5Z7Z
 
 | | Crystal structure of Striga hermonthica Dwarf14 (ShD14) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dwarf 14, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
5Z7W
 
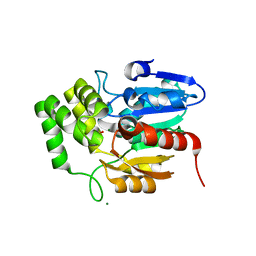 | | Crystal structure of Striga hermonthica HTL1 (ShHTL1) | | Descriptor: | GLYCEROL, Hyposensitive to light 1, MAGNESIUM ION, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
4TWD
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with memantine | | Descriptor: | Cys-loop ligand-gated ion channel, Memantine | | Authors: | Ulens, C, Spurny, R, Thompson, A.J, Alqazzaz, M, Debaveye, S, Lu, H, Price, K, Villalgordo, J.M, Tresadern, G, Lynch, J.W, Lummis, S.C.R. | | Deposit date: | 2014-06-30 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Prokaryote Ligand-Gated Ion Channel ELIC Captured in a Pore Blocker-Bound Conformation by the Alzheimer's Disease Drug Memantine.
Structure, 22, 2014
|
|
4TWH
 
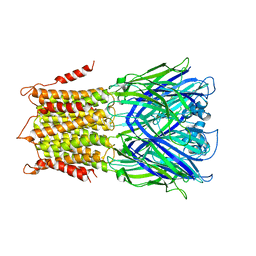 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) mutant F16'S | | Descriptor: | Cys-loop ligand-gated ion channel | | Authors: | Ulens, C, Spurny, R, Thompson, A.J, Alqazzaz, M, Debaveye, S, Lu, H, Price, K, Villalgordo, J.M, Tresadern, G, Lynch, J.W, Lummis, S.C.R. | | Deposit date: | 2014-06-30 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Prokaryote Ligand-Gated Ion Channel ELIC Captured in a Pore Blocker-Bound Conformation by the Alzheimer's Disease Drug Memantine.
Structure, 22, 2014
|
|
4TWF
 
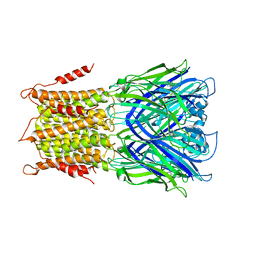 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromomemantine | | Descriptor: | Bromomemantine, Cys-loop ligand-gated ion channel | | Authors: | Ulens, C, Spurny, R, Thompson, A.J, Alqazzaz, M, Debaveye, S, Lu, H, Price, K, Villalgordo, J.M, Tresadern, G, Lynch, J.W, Lummis, S.C.R. | | Deposit date: | 2014-06-30 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.901 Å) | | Cite: | The Prokaryote Ligand-Gated Ion Channel ELIC Captured in a Pore Blocker-Bound Conformation by the Alzheimer's Disease Drug Memantine.
Structure, 22, 2014
|
|
1TAE
 
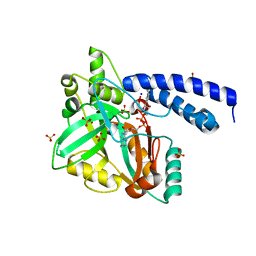 | |
1TA8
 
 | |
2HIV
 
 | | ATP-dependent DNA ligase from S. solfataricus | | Descriptor: | Thermostable DNA ligase | | Authors: | Pascal, J.M, Ellenberger, T. | | Deposit date: | 2006-06-29 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Flexible Interface between DNA Ligase and PCNA Supports Conformational Switching and Efficient Ligation of DNA.
Mol.Cell, 24, 2006
|
|
2HIX
 
 | |
7CR2
 
 | | human KCNQ2 in complex with retigabine | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 2, ethyl N-[2-azanyl-4-[(4-fluorophenyl)methylamino]phenyl]carbamate | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
3ZKR
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromoform | | Descriptor: | CYS-LOOP LIGAND-GATED ION CHANNEL, TRIBROMOMETHANE | | Authors: | Spurny, R, Billen, B, Howard, R.J, Brams, M, Debaveye, S, Price, K.L, Weston, D.A, Strelkov, S.V, Tytgat, J, Bertrand, S, Bertrand, D, Lummis, S.C.R, Ulens, C. | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.649 Å) | | Cite: | Multisite Binding of a General Anesthetic to the Prokaryotic Pentameric Erwinia Chrysanthemi Ligand-Gated Ion Channel (Elic).
J.Biol.Chem., 288, 2013
|
|
4XTX
 
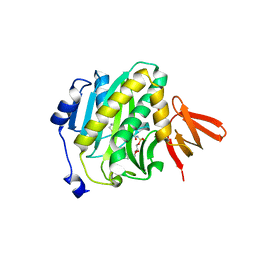 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor 57 with azide in place of ribose 2'OH | | Descriptor: | 1,2-ETHANEDIOL, 9-[2-azido-2-deoxy-5-O-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}sulfamoyl)-beta-D-arabinofuranosyl]-9H-purin-6-amine, Bifunctional ligase/repressor BirA | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.30010319 Å) | | Cite: | Targeting Mycobacterium tuberculosis Biotin Protein Ligase (MtBPL) with Nucleoside-Based Bisubstrate Adenylation Inhibitors.
J.Med.Chem., 58, 2015
|
|
4XTY
 
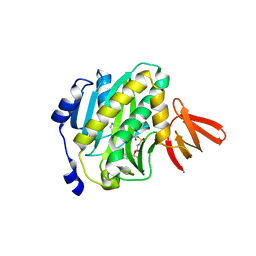 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor 63 with Fluorine in place of 2'OH | | Descriptor: | 2',5'-dideoxy-2'-fluoro-5'-[({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}sulfamoyl)amino]adenosine, Bifunctional ligase/repressor BirA | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.80002666 Å) | | Cite: | Targeting Mycobacterium tuberculosis Biotin Protein Ligase (MtBPL) with Nucleoside-Based Bisubstrate Adenylation Inhibitors.
J.Med.Chem., 58, 2015
|
|
3V8J
 
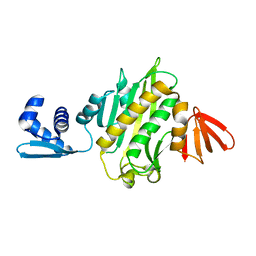 | |
4XTU
 
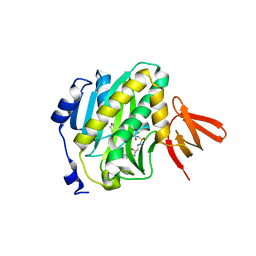 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor (N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide) | | Descriptor: | Bifunctional ligase/repressor BirA, N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6500299 Å) | | Cite: | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor (N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide)
To be Published
|
|
4XU2
 
 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor 87 with a 3'deoxy ribose | | Descriptor: | 3',5'-dideoxy-5'-[({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}sulfamoyl)amino]adenosine, Bifunctional ligase/repressor BirA | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8500284 Å) | | Cite: | Targeting Mycobacterium tuberculosis Biotin Protein Ligase (MtBPL) with Nucleoside-Based Bisubstrate Adenylation Inhibitors.
J.Med.Chem., 58, 2015
|
|
4XTW
 
 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor 46 with azide in place of 2'OH | | Descriptor: | 1,2-ETHANEDIOL, 2'-azido-2',5'-dideoxy-5'-[({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}sulfamoyl)amino]adenosine, Bifunctional ligase/repressor BirA | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.30014467 Å) | | Cite: | Targeting Mycobacterium tuberculosis Biotin Protein Ligase (MtBPL) with Nucleoside-Based Bisubstrate Adenylation Inhibitors.
J.Med.Chem., 58, 2015
|
|
4XTZ
 
 | |
4A98
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromoflurazepam | | Descriptor: | 7-BROMO-1-[2-(DIETHYLAMINO)ETHYL]-5-(2-FLUOROPHENYL)-1,3-DIHYDRO-2H-1,4-BENZODIAZEPIN-2-ONE, CYS-LOOP LIGAND-GATED ION CHANNEL | | Authors: | Spurny, R, Brams, M, Nury, H, Legrand, P, Ulens, C. | | Deposit date: | 2011-11-24 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Pentameric Ligand-Gated Ion Channel Elic is Activated by Gaba and Modulated by Benzodiazepines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4A97
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with zopiclone | | Descriptor: | (5R)-6-(5-chloropyridin-2-yl)-7-oxo-6,7-dihydro-5H-pyrrolo[3,4-b]pyrazin-5-yl 4-methylpiperazine-1-carboxylate, CYS-LOOP LIGAND-GATED ION CHANNEL | | Authors: | Spurny, R, Brams, M, Ulens, C. | | Deposit date: | 2011-11-24 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.343 Å) | | Cite: | Pentameric Ligand-Gated Ion Channel Elic is Activated by Gaba and Modulated by Benzodiazepines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4XU0
 
 | |
