8J2J
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, StayGold(Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2I
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (Q140S) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, StayGold(Q140S) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J3J
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with double mutations (Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, StayGold(Q140S, Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2L
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with double mutations (N137A, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2H
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (N137A) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | GLYCEROL, SODIUM ION, StayGold(N137A) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
7XSU
 
 | | Cardiac sodium channel in complex with LqhIII | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-like toxin Lqh3, ... | | Authors: | Jiang, D, Catterall, W.A. | | Deposit date: | 2022-05-15 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for Nav1.5 Opening Modulated by a Gating Modifier Toxin
To Be Published
|
|
8J0J
 
 | | AtSLAC1 8D mutant in closed state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8J1E
 
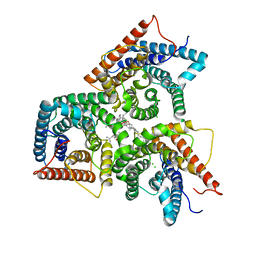 | | AtSLAC1 in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8C0T
 
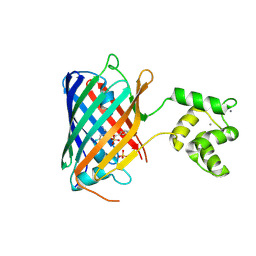 | | NRS 1.2: Fluorescent Sensors for Imaging Interstitial Calcium | | Descriptor: | CALCIUM ION, SULFATE ION, mNeonGreen,Optimized Ratiometric Calcium Sensor | | Authors: | Basquin, J, Griesbeck, O, Valiente-Gabioud, A. | | Deposit date: | 2022-12-19 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fluorescent sensors for imaging of interstitial calcium.
Nat Commun, 14, 2023
|
|
8I4J
 
 | |
7QLO
 
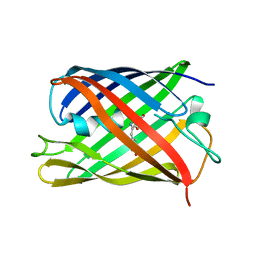 | | rsKiiro pump dump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLN
 
 | | rsKiiro pump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
8DN5
 
 | |
8DN2
 
 | |
8DN3
 
 | |
8DN4
 
 | | Cryo-EM structure of human Glycine Receptor alpha-1 beta heteromer, glycine-bound state3(desensitized state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-1, Glycine receptor subunit beta,Green fluorescent protein,Glycine receptor beta, ... | | Authors: | Liu, X, Wang, W. | | Deposit date: | 2022-07-10 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric gating of a human hetero-pentameric glycine receptor.
Nat Commun, 14, 2023
|
|
8GYF
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (K192Y) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, staygold(K192Y) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2022-09-22 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (K192Y) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8GOS
 
 | | Crystal structure of fluorescent protein RasM | | Descriptor: | RasM | | Authors: | Adachi, M, Kagotani, Y, Shimizu, R. | | Deposit date: | 2022-08-25 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Beat-frequency-resolved two-dimensional electronic spectroscopy: disentangling vibrational coherences in artificial fluorescent proteins with sub-10-fs visible laser pulses.
Opt Express, 31, 2023
|
|
7QLM
 
 | |
8SMU
 
 | | Integral fusion of the HtaA CR2 domain from Corynebacterium diphtheriae within EGFP | | Descriptor: | CHLORIDE ION, GLYCEROL, HtaACR2 integral fusion within enhanced green fluorescent protein, ... | | Authors: | Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2023-04-26 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development and atomic structure of a new fluorescence-based sensor to probe heme transfer in bacterial pathogens.
J.Inorg.Biochem., 249, 2023
|
|
8HCO
 
 | | Substrate-engaged TOM complex from yeast | | Descriptor: | Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, Mitochondrial import receptor subunit TOM5, ... | | Authors: | Zhou, X.Y, Yang, Y.Q, Wang, G.P, Wang, S.S. | | Deposit date: | 2022-11-02 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular pathway of mitochondrial preprotein import through the TOM-TIM23 supercomplex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YV3
 
 | |
7YRE
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, staygold(N137A,Q140S,Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
7YV5
 
 | |
8IMX
 
 | | Cryo-EM structure of GPI-T with a chimeric GPI-anchored protein | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, Y, Li, T, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
