5GRR
 
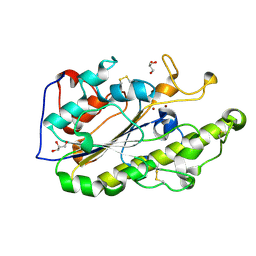 | | Crystal structure of MCR-1 | | Descriptor: | GLYCEROL, Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Ma, G, Zhu, Y, Yu, Z, Zhang, H. | | Deposit date: | 2016-08-12 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High resolution crystal structure of the catalytic domain of MCR-1
Sci Rep, 6, 2016
|
|
5FQL
 
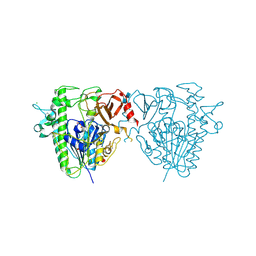 | | Insights into Hunter syndrome from the structure of iduronate-2- sulfatase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Demydchuk, M, Hill, C.H, Zhou, A, Bunkoczi, G, Stein, P.E, Marchesan, D, Deane, J.E, Read, R.J. | | Deposit date: | 2015-12-11 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into Hunter syndrome from the structure of iduronate-2-sulfatase.
Nat Commun, 8, 2017
|
|
2W5T
 
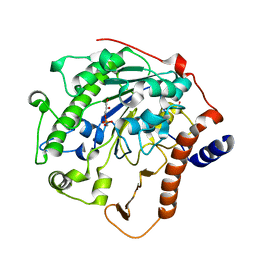 | | Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. | | Descriptor: | (2R)-2,3-dihydroxypropyl phosphate, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Lu, D, Wormann, M.E, Zhang, X, Schneewind, O, Grundling, A, Freemont, P.S. | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Mechanism of Lipoteichoic Acid Synthesis by Staphylococcus Aureus Ltas.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W5S
 
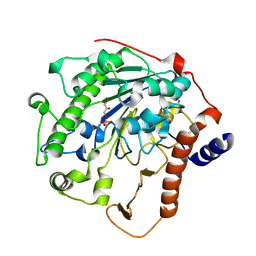 | | Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. | | Descriptor: | (2R)-2,3-dihydroxypropyl phosphate, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Lu, D, Wormann, M.E, Zhang, X, Schneewind, O, Grundling, A, Freemont, P.S. | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Mechanism of Lipoteichoic Acid Synthesis by Staphylococcus Aureus Ltas.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5GOV
 
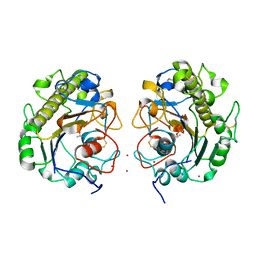 | | Crystal Structure of MCR-1, a phosphoethanolamine transferase, extracellular domain | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Hu, M, Guo, J, Chen, S, Hao, Q. | | Deposit date: | 2016-07-29 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of Escherichia coli originated MCR-1, a phosphoethanolamine transferase for Colistin Resistance.
Sci Rep, 6, 2016
|
|
7OZC
 
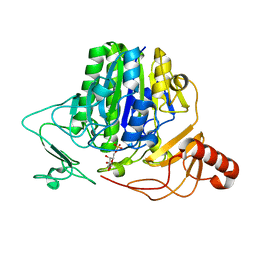 | |
7P26
 
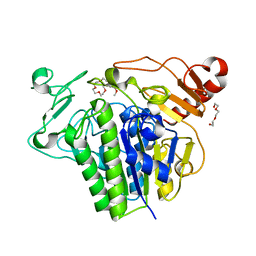 | |
7OZE
 
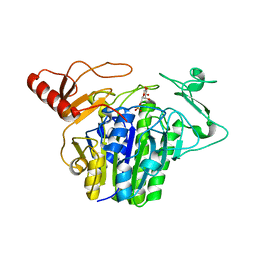 | |
5I5D
 
 | | Salmonella global domain 245 | | Descriptor: | Inner membrane protein YejM | | Authors: | Dong, C, Dong, H. | | Deposit date: | 2016-02-15 | | Release date: | 2017-04-19 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural insights into cardiolipin transfer from the Inner membrane to the outer membrane by PbgA in Gram-negative bacteria.
Sci Rep, 6, 2016
|
|
6XLP
 
 | | Structure of the essential inner membrane lipopolysaccharide-PbgA complex | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-deoxy-3-O-[(1R,3R)-1,3-dihydroxytetradecyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-1-O-phosphono-alpha-D-glucopyranose-(6-1)-[3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)]1,5-anhydro-2-deoxy-2-{[(1S,3R)-1-hydroxy-3-(pentanoyloxy)undecyl]amino}-4-O-phosphono-D-glucitol, ... | | Authors: | Payandeh, J, Clairefeuille, T. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the essential inner membrane lipopolysaccharide-PbgA complex.
Nature, 584, 2020
|
|
3B5Q
 
 | |
3ED4
 
 | | Crystal structure of putative arylsulfatase from escherichia coli | | Descriptor: | ARYLSULFATASE, GLYCEROL, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Gilmore, M, Chang, S, Bain, K, Wasserman, S, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Arylsulfatase from Escherichia Coli
To be Published
|
|
1AUK
 
 | | HUMAN ARYLSULFATASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARYLSULFATASE A, MAGNESIUM ION | | Authors: | Lukatela, G, Krauss, N, Theis, K, Gieselmann, V, Von Figura, K, Saenger, W. | | Deposit date: | 1997-08-29 | | Release date: | 1998-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human arylsulfatase A: the aldehyde function and the metal ion at the active site suggest a novel mechanism for sulfate ester hydrolysis.
Biochemistry, 37, 1998
|
|
1P49
 
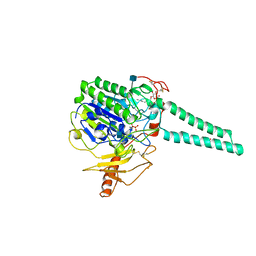 | | Structure of Human Placental Estrone/DHEA Sulfatase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Hernandez-Guzman, F.G, Higashiyama, T, Pangborn, W, Osawa, Y, Ghosh, D. | | Deposit date: | 2003-04-21 | | Release date: | 2003-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Human Estrone Sulfatase Suggests Functional Roles of Membrane Association
J.Biol.Chem., 278, 2003
|
|
2QZU
 
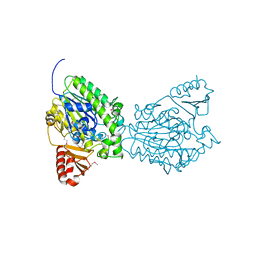 | | Crystal structure of the putative sulfatase yidJ from Bacteroides fragilis. Northeast Structural Genomics Consortium target BfR123 | | Descriptor: | Putative sulfatase yidJ | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Wang, D, Cunningham, K, Maglaqui, M, Owens, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-08-17 | | Release date: | 2007-09-04 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystal structure of the putative sulfatase yidJ from Bacteroides fragilis.
To be Published
|
|
8DI0
 
 | |
4MIV
 
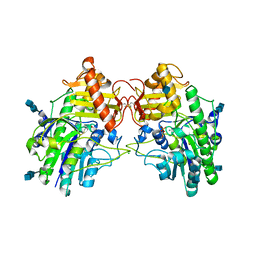 | | Crystal Structure of Sulfamidase, Crystal Form L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Sheldrick, G.M, Gaertner, J, Kraetzner, R, Steinfeld, R. | | Deposit date: | 2013-09-02 | | Release date: | 2014-05-14 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7LJ2
 
 | |
7LHA
 
 | |
4MHX
 
 | | Crystal Structure of Sulfamidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Gaertner, J, Kraetzner, R, Steinfeld, R, Sheldrick, G.M. | | Deposit date: | 2013-08-30 | | Release date: | 2014-05-14 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7PTH
 
 | |
7PTJ
 
 | |
8EG3
 
 | |
1N2L
 
 | | Crystal structure of a covalent intermediate of endogenous human arylsulfatase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARYLSULFATASE A, ... | | Authors: | Chruszcz, M, Laidler, P, Monkiewicz, M, Ortlund, E, Lebioda, L, Lewinski, K. | | Deposit date: | 2002-10-23 | | Release date: | 2003-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a covalent intermediate of endogenous human arylsulfatase A.
J.Inorg.Biochem., 96, 2003
|
|
1N2K
 
 | | Crystal structure of a covalent intermediate of endogenous human arylsulfatase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARYLSULFATASE A, ... | | Authors: | Chruszcz, M, Laidler, P, Monkiewicz, M, Ortlund, E, Lebioda, L, Lewinski, K. | | Deposit date: | 2002-10-23 | | Release date: | 2003-12-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of a covalent intermediate of endogenous human arylsulfatase A.
J.Inorg.Biochem., 96, 2003
|
|
