1M25
 
 | | STRUCTURE OF SYNTHETIC 26-MER PEPTIDE CONTAINING 145-169 SHEEP PRION PROTEIN SEGMENT AND C-TERMINAL CYSTEINE IN TFE SOLUTION | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Megy, S, Bertho, G, Kozin, S.A, Coadou, G, Debey, P, Hoa, G.H, Girault, J.-P. | | Deposit date: | 2002-06-21 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Possible role of region 152-156 in the structural duality of a peptide fragment from sheep prion protein
Protein Sci., 13, 2004
|
|
1MNL
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | Descriptor: | MONELLIN | | Authors: | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | Deposit date: | 1998-08-06 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
1M2J
 
 | | Sir2 homologue H80N mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
7QCA
 
 | | Spraguea lophii ribosome | | Descriptor: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S10, ... | | Authors: | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | Deposit date: | 2021-11-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state
Nat Microbiol, 2023
|
|
1MQJ
 
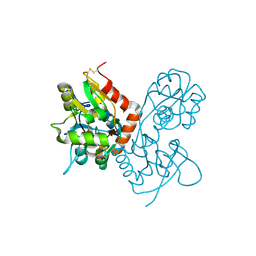 | | Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with willardiine at 1.65 angstroms resolution | | Descriptor: | 2-AMINO-3-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, ZINC ION, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1M3A
 
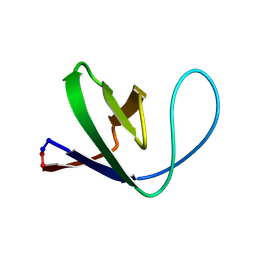 | | Solution structure of a circular form of the truncated N-terminal SH3 domain from oncogene protein c-Crk. | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
1M51
 
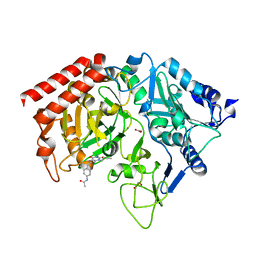 | | PEPCK complex with a GTP-competitive inhibitor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Foley, L.H, Wang, P, Dunten, P, Wertheimer, S.J. | | Deposit date: | 2002-07-06 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray Structures of two xanthine inhibitors bound to PEPCK and N-3 modifications of substituted 1,8-Dibenzylxanthines
Bioorg.Med.Chem.Lett., 13, 2003
|
|
7QB2
 
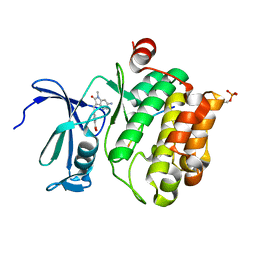 | | Pim1 in complex with (E)-4-((6-amino-1-methyl-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(E)-(6-azanyl-1-methyl-2-oxidanylidene-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
7QDU
 
 | |
1M3K
 
 | | biosynthetic thiolase, inactive C89A mutant | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-28 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
5CYS
 
 | |
1MJS
 
 | |
1M8O
 
 | | Platelet integrin alfaIIb-beta3 cytoplasmic domain | | Descriptor: | platelet integrin alfaIIb subunit: cytoplasmic domain, platelet integrin beta3 subunit: cytoplasmic domain | | Authors: | Vinogradova, O, Velyvis, A, Velyviene, A, Hu, B, Haas, T, Plow, E.F, Qin, J. | | Deposit date: | 2002-07-25 | | Release date: | 2002-09-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Structural mechanism of integrin alfaiib beta3
"Inside-out" Activation as regulated by its cytoplasmic face.
Cell(Cambridge,Mass.), 110, 2002
|
|
1MK8
 
 | | Crystal Structure of a Mutant Cytochrome c Peroxidase showing a Novel Trp-Tyr Covalent Cross-link | | Descriptor: | Cytochrome c Peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Immoos, C.E, Shimizu, H, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2002-08-28 | | Release date: | 2003-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Novel Heme and Peroxide-Dependent Tryptophan-Tyrosine Cross-Link in a Mutant of Cytochrome c Peroxidase
J.Mol.Biol., 328, 2003
|
|
5CZZ
 
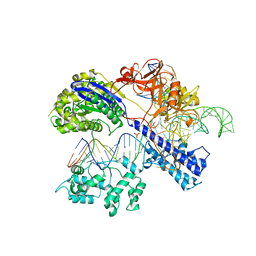 | | Crystal structure of Staphylococcus aureus Cas9 in complex with sgRNA and target DNA (TTGAAT PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Staphylococcus aureus Cas9.
Cell, 162, 2015
|
|
1MKN
 
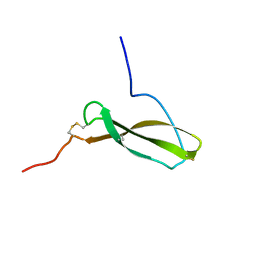 | | N-TERMINAL HALF OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
1ML3
 
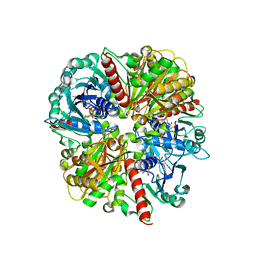 | | Evidences for a flip-flop catalytic mechanism of Trypanosoma cruzi glyceraldehyde-3-phosphate dehydrogenase, from its crystal structure in complex with reacted irreversible inhibitor 2-(2-phosphono-ethyl)-acrylic acid 4-nitro-phenyl ester | | Descriptor: | (3-FORMYL-BUT-3-ENYL)-PHOSPHONIC ACID, Glyceraldehyde 3-phosphate dehydrogenase, glycosomal, ... | | Authors: | Castilho, M.S, Pavao, F, Oliva, G. | | Deposit date: | 2002-08-29 | | Release date: | 2003-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence for the Two Phosphate Binding Sites of an Analogue of the Thioacyl Intermediate for the Trypanosoma cruzi Glyceraldehyde-3-phosphate Dehydrogenase-Catalyzed Reaction, from Its Crystal Structure.
Biochemistry, 42, 2003
|
|
7QFM
 
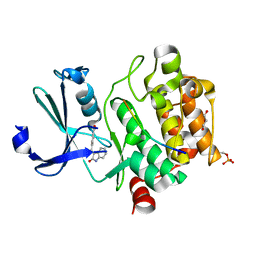 | | Pim1 in complex with (E)-4-((2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
1MM4
 
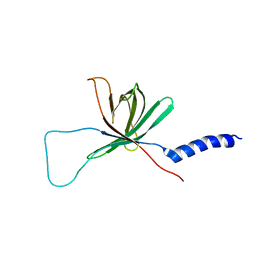 | | Solution NMR structure of the outer membrane enzyme PagP in DPC micelles | | Descriptor: | CrcA protein | | Authors: | Hwang, P.M, Choy, W.-Y, Lo, E.I, Chen, L, Forman-Kay, J.D, Raetz, C.R.H, Prive, G.G, Bishop, R.E, Kay, L.E. | | Deposit date: | 2002-09-03 | | Release date: | 2002-09-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Outer Membrane Enzyme PagP by NMR
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5VF1
 
 | |
7QFF
 
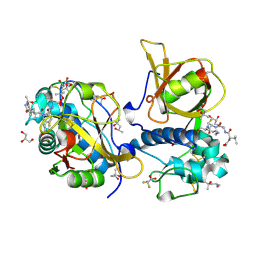 | | Peptide VACKSSQP in complex with human cathepsin V C25A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACE-VAL-ALA-CYS-LYS, CHLORIDE ION, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
4F5P
 
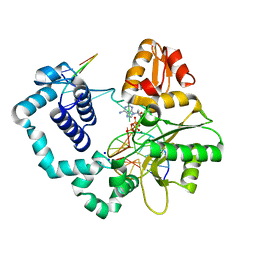 | | Open ternary mismatch complex of R283K DNA polymerase beta with a dATP analog | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Beard, W.A, Wilson, S.H. | | Deposit date: | 2012-05-13 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of dNTP Intermediate States during DNA Polymerase Active Site Assembly.
Structure, 20, 2012
|
|
7QG9
 
 | | Tail tip of siphophage T5 : common core proteins | | Descriptor: | Distal tail protein, L-shaped tail fiber protein p132, Minor tail protein, ... | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-21 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
1MDX
 
 | | Crystal structure of ArnB transferase with pyridoxal 5' phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, ArnB aminotransferase, GLYCEROL, ... | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T.A, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
1MP0
 
 | | Binary Complex of Human Glutathione-Dependent Formaldehyde Dehydrogenase with NAD(H) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Sanghani, P.C, Robinson, H, Hurley, T.D, Bosron, W.F. | | Deposit date: | 2002-09-10 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-function relationships in human Class III alcohol dehydrogenase
(formaldehyde dehydrogenase)
Chem.Biol.Interact., 143, 2003
|
|
