5VOF
 
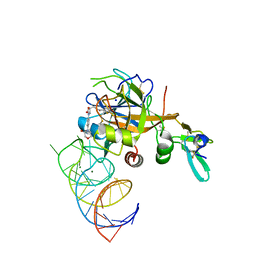 | | DesGla-XaS195A Bound to Aptamer 11F7t and Rivaroxaban | | Descriptor: | 5-chloro-N-({(5S)-2-oxo-3-[4-(3-oxomorpholin-4-yl)phenyl]-1,3-oxazolidin-5-yl}methyl)thiophene-2-carboxamide, CALCIUM ION, Coagulation factor X, ... | | Authors: | Krishnaswamy, S, Kumar, S. | | Deposit date: | 2017-05-02 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass.
Nat. Biotechnol., 36, 2018
|
|
6RTA
 
 | |
6TFH
 
 | |
5TDW
 
 | | Set3 PHD finger in complex with histone H3K4me3 | | Descriptor: | SET domain-containing protein 3, SODIUM ION, ZINC ION, ... | | Authors: | Andrews, F.H, Ali, M, Kutateladze, T.G. | | Deposit date: | 2016-09-19 | | Release date: | 2016-10-19 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight into Recognition of Methylated Histone H3K4 by Set3.
J. Mol. Biol., 429, 2017
|
|
5FFR
 
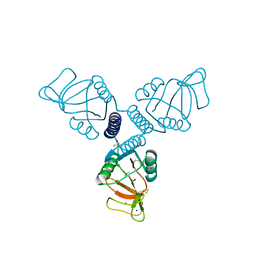 | | Crystal Structure of Surfactant Protein-A complexed with phosphocholine | | Descriptor: | CALCIUM ION, PHOSPHOCHOLINE, Pulmonary surfactant-associated protein A, ... | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
6H90
 
 | |
5FH3
 
 | |
6H83
 
 | | MamM CTD D249H - Copper form | | Descriptor: | BETA-MERCAPTOETHANOL, COPPER (II) ION, Magnetosome protein MamM, ... | | Authors: | Barber-Zucker, S, Zarivach, R. | | Deposit date: | 2018-08-01 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | MamM CTD D249H - Copper form
To Be Published
|
|
5T9B
 
 | |
6TLS
 
 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 4,6-DIBROMOBENZOTRIAZOLE | | Descriptor: | 5,7-bis(bromanyl)-1~{H}-benzotriazole, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
1HDA
 
 | |
6TK2
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 1ms structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, SODIUM ION, ... | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6TLL
 
 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 4,5,6,7-TETRABROMOBENZOTRIAZOLE (tBBT) | | Descriptor: | 4,5,6,7-TETRABROMOBENZOTRIAZOLE, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
5HI0
 
 | | The Substrate Binding Mode and Chemical Basis of a Reaction Specificity Switch in Oxalate Decarboxylase | | Descriptor: | COBALT (II) ION, OXALATE ION, Oxalate decarboxylase OxdC, ... | | Authors: | Zhu, W, Easthon, L.M, Reinhardt, L.A, Tu, C, Cohen, S.E, Silverman, D.N, Allen, K.N, Richards, N.G.J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Substrate Binding Mode and Molecular Basis of a Specificity Switch in Oxalate Decarboxylase.
Biochemistry, 55, 2016
|
|
5TNZ
 
 | | HtrA2 S142D mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SODIUM ION, Serine protease HTRA2, ... | | Authors: | Macedo-Ribeiro, S, Merski, M, Pereira, P.J.B. | | Deposit date: | 2016-10-15 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular motion regulates the activity of the Mitochondrial Serine Protease HtrA2.
Cell Death Dis, 8, 2017
|
|
6THV
 
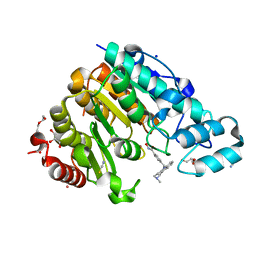 | | X-ray structure of the Danio rerio histone deacetylase 6 (HDAC6; catalytic domain 2) in complex with Tubastatin A | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-methyl-3,4-dihydro-1~{H}-pyrido[4,3-b]indol-5-yl)methyl]-~{N}-oxidanyl-benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Barinka, C, Motlova, L, Svoboda, M. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and in Vivo Characterization of Tubastatin A, a Widely Used Histone Deacetylase 6 Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
6HON
 
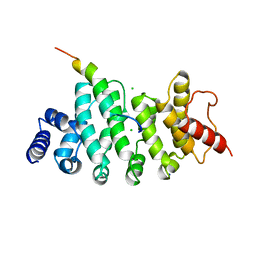 | | Drosophila NOT4 CBM peptide bound to human CAF40 | | Descriptor: | CCR4-NOT transcription complex subunit 4, isoform L, CCR4-NOT transcription complex subunit 9, ... | | Authors: | Raisch, T, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A conserved CAF40-binding motif in metazoan NOT4 mediates association with the CCR4-NOT complex.
Genes Dev., 33, 2019
|
|
5E1I
 
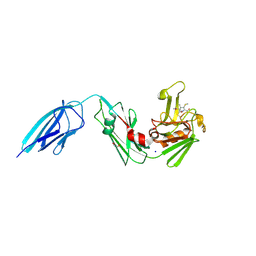 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T210 | | Descriptor: | (2S,3R,4R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-(methylsulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, GLYCEROL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-29 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
2CKS
 
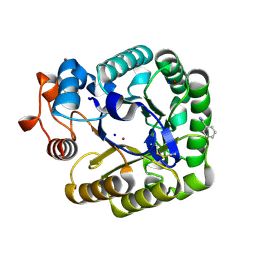 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THERMOBIFIDA FUSCA ENDOGLUCANASE CEL5A (E5) | | Descriptor: | BENZAMIDINE, ENDOGLUCANASE E-5, SODIUM ION, ... | | Authors: | Berglund, G.I, Gualfetti, P.J, Requadt, C, Gross, L.S, Bergfors, T, Shaw, A, Saldajeno, M, Mitchinson, C, Sandgren, M. | | Deposit date: | 2006-04-21 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Catalytic Domain of Thermobifida Fusca Endoglucanase Cel5A in Complex with Cellotetraose
To be Published
|
|
5DMU
 
 | | Structure of the NHEJ polymerase from Methanocella paludicola | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NHEJ Polymerase, ... | | Authors: | Brissett, N.C, Bartlett, E.J, Doherty, A.J. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Molecular basis for DNA strand displacement by NHEJ repair polymerases.
Nucleic Acids Res., 44, 2016
|
|
6TK1
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 20ms structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, SODIUM ION, ... | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
5T91
 
 | | Crystal structure of B. subtilis 168 GlpQ in complex with bicine | | Descriptor: | BICINE, CALCIUM ION, Glycerophosphoryl diester phosphodiesterase, ... | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Identification of Two Phosphate Starvation-induced Wall Teichoic Acid Hydrolases Provides First Insights into the Degradative Pathway of a Key Bacterial Cell Wall Component.
J. Biol. Chem., 291, 2016
|
|
6TQP
 
 | | Structural insight into tanapoxvirus mediated inhibition of apoptosis | | Descriptor: | 16L protein, Bcl-2-binding component 3, isoforms 1/2, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2019-12-17 | | Release date: | 2020-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84940946 Å) | | Cite: | Structural insight into tanapoxvirus-mediated inhibition of apoptosis.
Febs J., 287, 2020
|
|
6HHD
 
 | | Mouse Prion Protein in complex with Nanobody 484 | | Descriptor: | CHLORIDE ION, GLYCEROL, Major prion protein, ... | | Authors: | Soror, S, Abskharon, R, Wohlkonig, A. | | Deposit date: | 2018-08-28 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion.
Plos Pathog., 15, 2019
|
|
5HKY
 
 | | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase CBL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form)
To be published
|
|
