1G3U
 
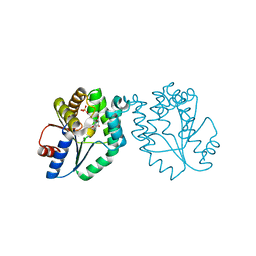 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE MONOPHOSPHATE (TMP) | | Descriptor: | MAGNESIUM ION, SULFATE ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Li de la Sierra, I, Munier-Lehmann, H, Gilles, A.M, Barzu, O, Delarue, M. | | Deposit date: | 2000-10-25 | | Release date: | 2001-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray structure of TMP kinase from Mycobacterium tuberculosis complexed with TMP at 1.95 A resolution.
J.Mol.Biol., 311, 2001
|
|
1G3V
 
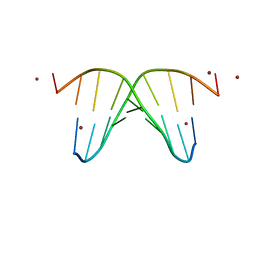 | | CRYSTAL STRUCTURE OF NICKEL-D[CGTGTACACG]2 | | Descriptor: | 5'-D(*CP*GP*TP*GP*TP*AP*CP*AP*CP*G)-3', NICKEL (II) ION | | Authors: | Subirana, J.A, Abrescia, N.G.A. | | Deposit date: | 2000-10-25 | | Release date: | 2002-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nickel-guanine interactions in DNA: crystal structure of nickel-d[CGTGTACACG]2.
J.Biol.Inorg.Chem., 7, 2002
|
|
1G3W
 
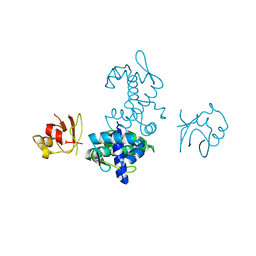 | | CD-CYS102SER DTXR | | Descriptor: | CADMIUM ION, DIPHTHERIA TOXIN REPRESSOR, SULFATE ION | | Authors: | Pohl, E, Goranson-Siekierke, J, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2000-10-25 | | Release date: | 2001-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of three diphtheria toxin repressor (DtxR) variants with decreased repressor activity.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G3X
 
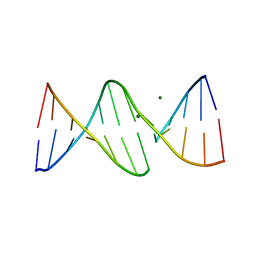 | | INTERCALATION OF AN 9ACRIDINE-PEPTIDE DRUG IN A DNA DODECAMER | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION, N(ALPHA)-(9-ACRIDINOYL)-TETRAARGININE-AMIDE | | Authors: | Malinina, L, Soler-Lopez, M, Aymami, J, Subirana, J.A. | | Deposit date: | 2000-10-25 | | Release date: | 2002-08-16 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Intercalation of an acridine-peptide drug in an AA/TT base step in the crystal structure of [d(CGCGAATTCGCG)](2) with six duplexes and seven Mg(2+) ions in the asymmetric unit.
Biochemistry, 41, 2002
|
|
1G3Y
 
 | | ARG80ALA DTXR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR, ZINC ION | | Authors: | Pohl, E, Goranson-Siekierke, J, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2000-10-25 | | Release date: | 2001-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of three diphtheria toxin repressor (DtxR) variants with decreased repressor activity.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G3Z
 
 | | CARBONIC ANHYDRASE II (F131V) | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, ZINC ION | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-25 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Contribution of Fluorine to Protein-Ligand Affinity in the Binding of Fluoroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
1G41
 
 | | CRYSTAL STRUCTURE OF HSLU HAEMOPHILUS INFLUENZAE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK PROTEIN HSLU, SULFATE ION | | Authors: | Trame, C.B, McKay, D.B. | | Deposit date: | 2000-10-25 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Haemophilus influenzae HslU protein in crystals with one-dimensional disorder twinning.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G42
 
 | | STRUCTURE OF 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE (LINB) FROM SPHINGOMONAS PAUCIMOBILIS COMPLEXED WITH 1,2-DICHLOROPROPANE | | Descriptor: | 1,2-DICHLORO-PROPANE, 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE, ACETATE ION, ... | | Authors: | Oakley, A.J, Prokop, Z, Bohac, M, Kmunicek, J, Jedlicka, T, Monincova, M, Kuta-Smatanova, I, Nagata, Y, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2000-10-26 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition.
Biochemistry, 41, 2002
|
|
1G43
 
 | | CRYSTAL STRUCTURE OF A FAMILY IIIA CBD FROM CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | CALCIUM ION, SCAFFOLDING PROTEIN, ZINC ION | | Authors: | Shimon, L.J.W, Pages, S, Belaich, A, Belaich, J.-P, Bayer, E.A, Lamed, R, Shoham, Y, Frolow, F. | | Deposit date: | 2000-10-26 | | Release date: | 2000-12-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a family IIIa scaffoldin CBD from the cellulosome of Clostridium cellulolyticum at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1G45
 
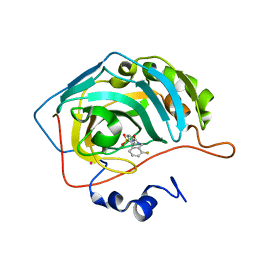 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2-FLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-(2-FLUORO-BENZYL)-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-26 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Contribution of Flourine to Protein-ligand Affinity in the Binding of Flouroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
1G46
 
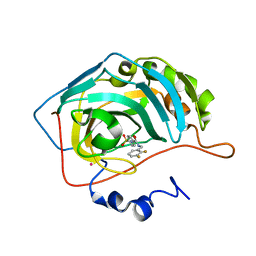 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,3-DIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-(2,3-DIFLUORO-BENZYL)-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-26 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Contribution of Flourine to Protein-ligand Affinity in the Binding of Flouroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
1G47
 
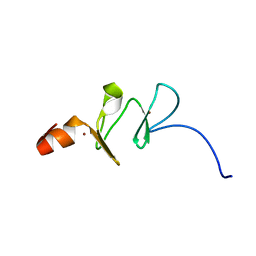 | | 1ST LIM DOMAIN OF PINCH PROTEIN | | Descriptor: | PINCH PROTEIN, ZINC ION | | Authors: | Velyvis, A, Yang, Y, Wu, C, Qin, J. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the focal adhesion adaptor PINCH LIM1 domain and characterization of its interaction with the integrin-linked kinase ankyrin repeat domain.
J.Biol.Chem., 276, 2001
|
|
1G48
 
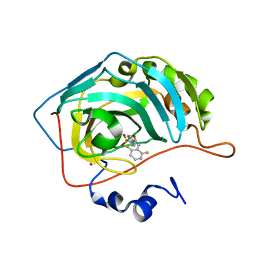 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,6-DIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-(2,6-DIFLUORO-BENZYL)-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-26 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Contribution of Flourine to Protein-ligand Affinity in the Binding of Flouroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
1G49
 
 | | A CARBOXYLIC ACID BASED INHIBITOR IN COMPLEX WITH MMP3 | | Descriptor: | (1N)-4-N-BUTOXYPHENYLSULFONYL-(2R)-N-HYDROXYCARBOXAMIDO-(4S)-METHANESULFONYLAMINO-PYRROLIDINE, CALCIUM ION, MATRIX METALLOPROTEINASE 3, ... | | Authors: | Natchus, M.G, Bookland, R.G, De, B, Almstead, N.G, Pikul, S. | | Deposit date: | 2000-10-26 | | Release date: | 2001-10-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of new hydroxamate matrix metalloproteinase inhibitors derived from functionalized 4-aminoprolines.
J.Med.Chem., 43, 2000
|
|
1G4A
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
1G4B
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
1G4C
 
 | |
1G4D
 
 | |
1G4E
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | THIAMIN PHOSPHATE SYNTHASE | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-10-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
1G4F
 
 | | NMR STRUCTURE OF THE FIFTH DOMAIN OF HUMAN BETA2-GLYCOPROTEIN I | | Descriptor: | BETA2-GLYCOPROTEIN I | | Authors: | Hoshino, M, Hagihara, Y, Nishii, I, Yamazaki, T, Kato, H, Goto, Y. | | Deposit date: | 2000-10-27 | | Release date: | 2000-11-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Identification of the phospholipid-binding site of human beta(2)-glycoprotein I domain V by heteronuclear magnetic resonance.
J.Mol.Biol., 304, 2000
|
|
1G4G
 
 | | NMR STRUCTURE OF THE FIFTH DOMAIN OF HUMAN BETA2-GLYCOPROTEIN I | | Descriptor: | BETA2-GLYCOPROTEIN I | | Authors: | Hoshino, M, Hagihara, Y, Nishii, I, Yamazaki, T, Kato, H, Goto, Y. | | Deposit date: | 2000-10-27 | | Release date: | 2000-11-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Identification of the phospholipid-binding site of human beta(2)-glycoprotein I domain V by heteronuclear magnetic resonance.
J.Mol.Biol., 304, 2000
|
|
1G4H
 
 | | LINB COMPLEXED WITH BUTAN-1-OL | | Descriptor: | 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE, 1-BUTANOL, CALCIUM ION, ... | | Authors: | Oakley, A.J, Prokop, Z, Bohac, M, Kmunicek, J, Jedlicka, T, Monincova, M, Kuta-Smatanova, I, Nagata, Y, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition.
Biochemistry, 41, 2002
|
|
1G4I
 
 | | Crystal structure of the bovine pancreatic phospholipase A2 at 0.97A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Steiner, R.A, Rozeboom, H.J, de Vries, A, Kalk, K.H, Murshudov, G.N, Wilson, K.S, Dijkstra, B.W. | | Deposit date: | 2000-10-27 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | X-ray structure of bovine pancreatic phospholipase A2 at atomic resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G4J
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,3,4,5,6-PENTAFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-(2,3,4,5,6-PENTAFLUORO-BENZYL)-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-27 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Contribution of Flourine to Protein-ligand Affinity in the Binding of Flouroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
1G4K
 
 | | X-ray Structure of a Novel Matrix Metalloproteinase Inhibitor Complexed to Stromelysin | | Descriptor: | 5-METHYL-5-(4-PHENOXY-PHENYL)-PYRIMIDINE-2,4,6-TRIONE, CALCIUM ION, GLYCEROL, ... | | Authors: | Dunten, P, Kammlott, U, Crowther, R, Levin, W, Foley, L.H, Wang, P, Palermo, R. | | Deposit date: | 2000-10-27 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of a novel matrix metalloproteinase inhibitor complexed to stromelysin.
Protein Sci., 10, 2001
|
|
