2LJ0
 
 | | The third SH3 domain of R85FL | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2011-09-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Basis for the Recognition of Ataxin-7 PRR with R85FL SH3 Domain
To be Published
|
|
4RKY
 
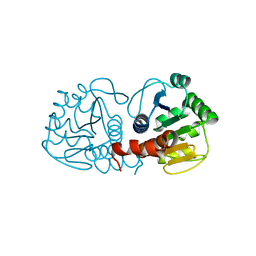 | | Crystal structure of DJ-1 isoform X1 | | Descriptor: | Protein DJ-1 | | Authors: | Liddington, R.C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in parkinson's disease models.
J.Neurosci., 34, 2014
|
|
6PEB
 
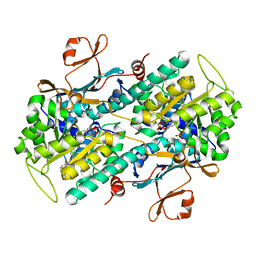 | | Crystal Structure of human NAMPT in complex with NVP-LTM976 | | Descriptor: | N-{4-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-3-(pyridin-3-yl)azetidine-1-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Weihofen, W.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Scaffold Morphing Identifies 3-Pyridyl Azetidine Ureas as Inhibitors of Nicotinamide Phosphoribosyltransferase (NAMPT).
Acs Med.Chem.Lett., 10, 2019
|
|
4RN3
 
 | |
4G1M
 
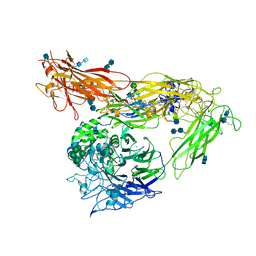 | | Re-refinement of alpha V beta 3 structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Springer, T.A, Mi, L, Zhu, J. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Alpha V Beta 3 Integrin Crystal Structures and their Functional Implications
Biochemistry, 51, 2012
|
|
4WUC
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM NaCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4S34
 
 | | ERK2 (I84A) in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Livnah, O, Karamansha, Y, Tzarum, N. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiple mechanisms render Erk proteins MEK-independent
To be Published
|
|
4WFL
 
 | |
6RNK
 
 | | Crystal structure of a humanized (K18E, K269N) rat succinate receptor SUCNR1 (GPR91) in complex with a nanobody and antagonist NF-56-EJ40. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[2-[[3-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, GLYCEROL, ... | | Authors: | Haffke, M, Jaakola, V.-P. | | Deposit date: | 2019-05-08 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis of species-selective antagonist binding to the succinate receptor.
Nature, 574, 2019
|
|
6P8G
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
4WIO
 
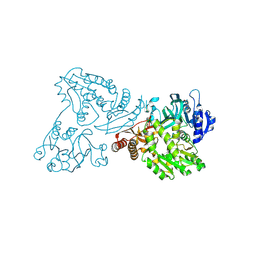 | | Crystal structure of the C89A GMP synthetase inactive mutant from Plasmodium falciparum in complex with glutamine | | Descriptor: | GLUTAMINE, GMP synthetase | | Authors: | Ballut, L, Violot, S, Haser, R, Aghajari, N. | | Deposit date: | 2014-09-26 | | Release date: | 2015-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Active site coupling in Plasmodium falciparum GMP synthetase is triggered by domain rotation.
Nat Commun, 6, 2015
|
|
6RFS
 
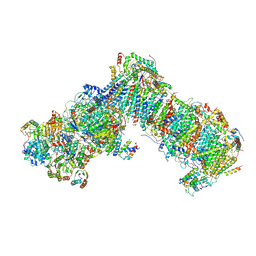 | | Cryo-EM structure of a respiratory complex I mutant lacking NDUFS4 | | Descriptor: | Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), Acyl carrier protein ACPM2 of NADH:Ubiquinone Oxidoreductase (Complex I), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Parey, K, Vonck, J. | | Deposit date: | 2019-04-16 | | Release date: | 2019-12-11 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | High-resolution cryo-EM structures of respiratory complex I: Mechanism, assembly, and disease.
Sci Adv, 5, 2019
|
|
4RM8
 
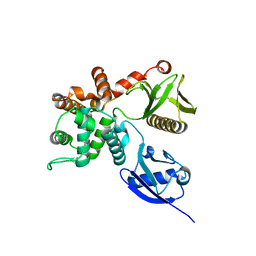 | | Crystal structure of human ezrin in space group P21 | | Descriptor: | Ezrin | | Authors: | Phang, J.M, Harrop, S.J, Davies, R, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2014-10-20 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin.
Biochem. J., 473, 2016
|
|
6RS8
 
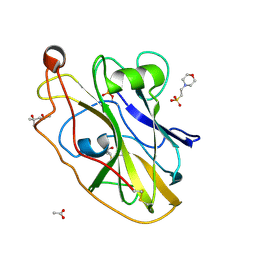 | | X-ray crystal structure of LsAA9B (transition metals soak) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
4WOI
 
 | | 4,5-linked aminoglycoside antibiotics regulate the bacterial ribosome by targeting dynamic conformational processes within intersubunit bridge B2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pulk, A, Cate, J.H.D, Blanchard, S, Wasserman, M, Altman, R, Zhou, Z, Zinder, J, Green, K, Garneau-Tsodikova, S. | | Deposit date: | 2014-10-15 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemically related 4,5-linked aminoglycoside antibiotics drive subunit rotation in opposite directions.
Nat Commun, 6, 2015
|
|
2LZP
 
 | |
4WWW
 
 | | Crystal structure of the E. coli ribosome bound to CEM-101 | | Descriptor: | (3aS,4R,7S,9R,10R,11R,13R,15R,15aR)-1-{4-[4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl]butyl}-4-ethyl-7-fluoro-11-methoxy-3a ,7,9,11,13,15-hexamethyl-2,6,8,14-tetraoxotetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-10-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S rRNA, 23S rRNA, ... | | Authors: | Dunkle, J.A, Zhang, W, Cate, J.H.D, Mankin, A.S. | | Deposit date: | 2014-11-12 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis.
Antimicrob. Agents Chemother., 54, 2010
|
|
4RVO
 
 | |
4WF1
 
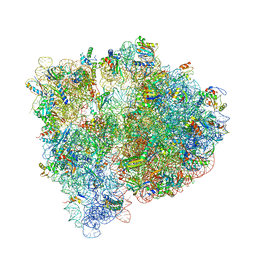 | | Crystal structure of the E. coli ribosome bound to negamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Olivier, N.B, Altman, R.B, Noeske, J, Basarab, G.S, Code, E, Ferguson, A.D, Gao, N, Huang, J, Juette, M.F, Livchak, S, Miller, M.D, Prince, D.B, Cate, J.H.D, Buurman, E.T, Blanchard, S.C. | | Deposit date: | 2014-09-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4S32
 
 | |
4WQ6
 
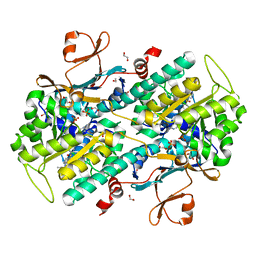 | | The crystal structure of human Nicotinamide phosphoribosyltransferase (NAMPT) in complex with N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide inhibitor (compound 21) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Li, D, Wang, W. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Identification of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with no evidence of CYP3A4 time-dependent inhibition and improved aqueous solubility.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6P8E
 
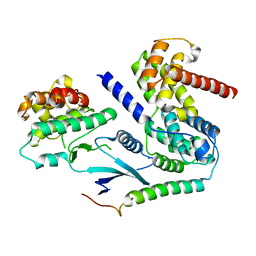 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1, ... | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
6PPU
 
 | | Cryo-EM structure of AdnAB-AMPPNP-DNA complex | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), DNA (29-MER), IRON/SULFUR CLUSTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4X1H
 
 | | Opsin/G(alpha) peptide complex stabilized by nonyl-glucoside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-terminal derived peptide of guanine nucleotide-binding protein G(t) subunit alpha-1, PALMITIC ACID, ... | | Authors: | Blankenship, E, Lodowski, D.T. | | Deposit date: | 2014-11-24 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The High-Resolution Structure of Activated Opsin Reveals a Conserved Solvent Network in the Transmembrane Region Essential for Activation.
Structure, 23, 2015
|
|
4X5R
 
 | | Crystal structure of FimH in complex with a squaryl-phenyl alpha-D-mannopyranoside derivative | | Descriptor: | 2-chloro-4-{[2-(4-methylpiperazin-1-yl)-3,4-dioxocyclobut-1-en-1-yl]amino}phenyl alpha-D-mannopyranoside, Protein FimH, SULFATE ION | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
