3MKV
 
 | | Crystal structure of amidohydrolase eaj56179 | | Descriptor: | CARBONATE ION, GLYCEROL, PUTATIVE AMIDOHYDROLASE, ... | | Authors: | Patskovsky, Y, Bonanno, J, Ozyurt, S, Sauder, J.M, Freeman, J, Wu, B, Smith, D, Bain, K, Rodgers, L, Wasserman, S.R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-15 | | Release date: | 2010-04-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3MKW
 
 | | Structure of sopB(155-272)-18mer complex, I23 form | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | Authors: | Schumacher, M.A, Piro, K, Xu, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3MKY
 
 | | Structure of SopB(155-323)-18mer DNA complex, I23 form | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | Authors: | Schumacher, M.A, Piro, K, Xu, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3MKZ
 
 | | Structure of SopB(155-272)-18mer complex, P21 form | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB | | Authors: | Schumacher, M.A. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3ML0
 
 | | Thermostable Penicillin G acylase from Alcaligenes faecalis in tetragonal form | | Descriptor: | CALCIUM ION, Penicillin G acylase, alpha subunit, ... | | Authors: | Varshney, N.K, Kumar, R.S, Ignatova, Z, Dodson, E, Suresh, C.G. | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and X-ray structure analysis of a thermostable penicillin G acylase from Alcaligenes faecalis.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3ML1
 
 | | Crystal Structure of the Periplasmic Nitrate Reductase from Cupriavidus necator | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DIOXOTHIOMOLYBDENUM(VI) ION, Diheme cytochrome c napB, ... | | Authors: | Coelho, C, Trincao, J, Romao, M.J. | | Deposit date: | 2010-04-16 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Cupriavidus necator nitrate reductase in oxidized and partially reduced states
J.Mol.Biol., 408, 2011
|
|
3ML2
 
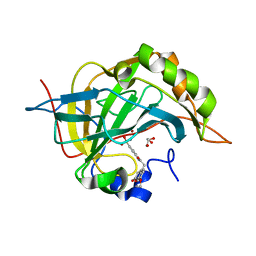 | | Human carbonic anhydsase II in complex with an aryl sulfonamide inhibitor | | Descriptor: | 2-(7-methoxy-2-oxo-2H-chromen-4-yl)-N-(4-sulfamoylphenyl)acetamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Avvaru, B.S, Wagner, J, Robbins, A.H, Mckenna, R. | | Deposit date: | 2010-04-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coumarinyl-substituted sulfonamides strongly inhibit several human carbonic anhydrase isoforms: solution and crystallographic investigations.
Bioorg.Med.Chem., 18, 2010
|
|
3ML3
 
 | | Crystal structure of the IcsA autochaperone region | | Descriptor: | Outer membrane protein icsA autotransporter | | Authors: | Diezmann, D, Kuhnel, K. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Autochaperone Region from the Shigella flexneri Autotransporter IcsA
J.Bacteriol., 193, 2011
|
|
3ML4
 
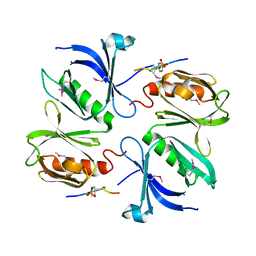 | |
3ML5
 
 | | Crystal structure of the C183S/C217S mutant of human CA VII in complex with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 7, ZINC ION | | Authors: | Di Fiore, A, Truppo, E, Supuran, C.T, Alterio, V, Dathan, N, Bootorabi, F, Parkkila, S, Monti, S.M, De Simone, G. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the C183S/C217S mutant of human CA VII in complex with acetazolamide
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3ML6
 
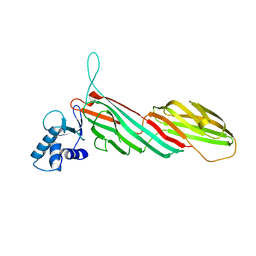 | | a complex between Dishevelled2 and clathrin adaptor AP-2 | | Descriptor: | Chimeric complex between protein Dishevelled2 homolog dvl-2 and clathrin adaptor AP-2 complex subunit mu | | Authors: | Yu, A, Xing, Y, Harrison, S.C, Kirchhausen, T.L. | | Deposit date: | 2010-04-16 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural analysis of the interaction between Dishevelled2 and clathrin AP-2 adaptor, a critical step in noncanonical Wnt signaling.
Structure, 18, 2010
|
|
3ML8
 
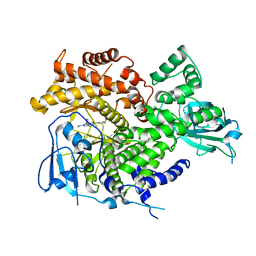 | |
3ML9
 
 | |
3MLA
 
 | | BaNadD in complex with inhibitor 1_02 | | Descriptor: | 4-[2-(anthracen-9-ylmethylidene)hydrazino]-N-(3-chlorophenyl)-4-oxobutanamide, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Huang, N, Eyobo, Y, Zhang, H. | | Deposit date: | 2010-04-16 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexes of bacterial nicotinate mononucleotide adenylyltransferase with inhibitors: implication for structure-based drug design and improvement.
J.Med.Chem., 53, 2010
|
|
3MLB
 
 | | BaNadD in complex with inhibitor 1_02_1 | | Descriptor: | 4,4'-{cyclohexa-2,5-diene-1,4-diylidenebis[(E)methylylidene(E)diazene-2,1-diyl]}bis[N-(2-chlorophenyl)-4-oxobutanamide], FORMIC ACID, POTASSIUM ION, ... | | Authors: | Huang, N, Zhang, H, Eyobo, Y. | | Deposit date: | 2010-04-16 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complexes of bacterial nicotinate mononucleotide adenylyltransferase with inhibitors: implication for structure-based drug design and improvement.
J.Med.Chem., 53, 2010
|
|
3MLC
 
 | | Crystal structure of FG41MSAD inactivated by 3-chloropropiolate | | Descriptor: | 3-chloro-3-oxopropanoic acid, FG41 Malonate Semialdehyde Decarboxylase | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-16 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
3MLE
 
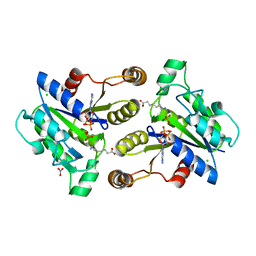 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori cocrystallized with ATP | | Descriptor: | 8-aminooctanoic acid, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nicholls, R, Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3MLF
 
 | |
3MLG
 
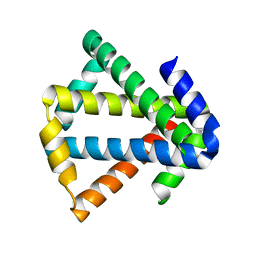 | | 2ouf-2x, a designed knotted protein | | Descriptor: | 2X chimera of Helicobacter pylori protein HP0242 | | Authors: | King, N.P, Sawaya, M.R, Jacobitz, A.W, Yeates, T.O. | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure and folding of a designed knotted protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MLH
 
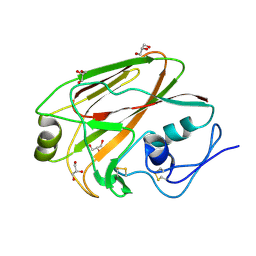 | | Crystal structure of the 2009 H1N1 influenza virus hemagglutinin receptor-binding domain | | Descriptor: | GLYCEROL, Hemagglutinin | | Authors: | DuBois, R.M, Aguilar-Yanez, J.M, Mendoza-Ochoa, G.I, Schultz-Cherry, S, Alvarez, M.M, White, S.W, Russell, C.J. | | Deposit date: | 2010-04-16 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Receptor-Binding Domain of Influenza Virus Hemagglutinin Produced in Escherichia coli Folds into Its Native, Immunogenic Structure.
J.Virol., 85, 2011
|
|
3MLI
 
 | | 2ouf-ds, a disulfide-linked dimer of Helicobacter pylori protein HP0242 | | Descriptor: | CALCIUM ION, Putative uncharacterized protein | | Authors: | King, N.P, Sawaya, M.R, Jacobitz, A.W, Yeates, T.O. | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and folding of a designed knotted protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MLJ
 
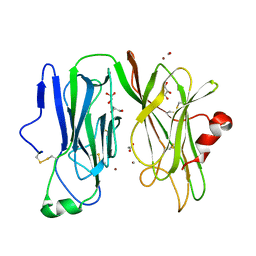 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound carbon monooxide (CO) | | Descriptor: | ACETATE ION, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Prigge, S.T, Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MLK
 
 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite | | Descriptor: | COPPER (II) ION, NICKEL (II) ION, NITRITE ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MLL
 
 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide | | Descriptor: | AZIDE ION, COPPER (II) ION, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MLM
 
 | | Crystal structure of Bn IV in complex with myristic acid: A Lys49 myotoxic phospholipase A2 from Bothrops neuwiedi venom | | Descriptor: | BN-IV Lys-49 Phospholipase A2, MYRISTIC ACID, SULFATE ION | | Authors: | Delatorre, P, Rocha, B.A.M, Cavada, B.S, Toyama, M.H, Toyama, D, Gadelha, C.A.A. | | Deposit date: | 2010-04-17 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of Bn IV in complex with myristic acid: a Lys49 myotoxic phospholipase A2 from Bothrops neuwiedi venom.
Biochimie, 93, 2011
|
|
