8AI2
 
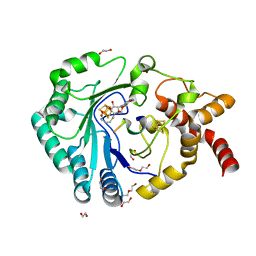 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI1
 
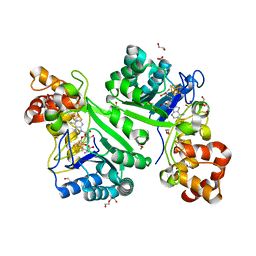 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-homocysteine bound. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Polsinelli, I, Chavas, L.M.G, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI0
 
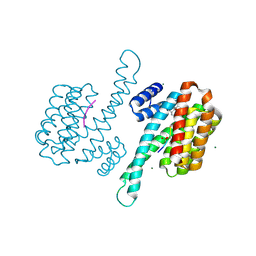 | | Small molecular stabilizer for ERalpha and 14-3-3sigma (1080268) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-(2-methyl-2-phenylazanyl-propanoyl)piperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AHZ
 
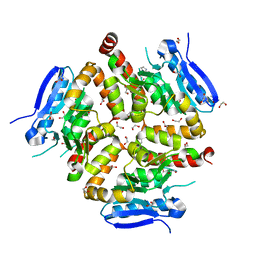 | | Native VirD of Streptomyces virginiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Enoyl-CoA hydratase, ... | | Authors: | Collin, S, Gruez, A. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8AHY
 
 | |
8AHW
 
 | | Structure of DCS-resistant variant D322N of alanine racemase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Alanine racemase, GLYCEROL | | Authors: | de Chiara, C, Prosser, G, Ogrodowicz, R.W, de Carvalho, L.P.S. | | Deposit date: | 2022-07-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of the d-Cycloserine-Resistant Variant D322N of Alanine Racemase from Mycobacterium tuberculosis .
Acs Bio Med Chem Au, 3, 2023
|
|
8AHU
 
 | | Crystal structure of D-amino acid aminotrensferase from Haliscomenobacter hydrossis complexed with D-cycloserine | | Descriptor: | Aminotransferase class IV, GLYCEROL, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Mechanism of D-Cycloserine Inhibition of D-Amino Acid Transaminase from Haliscomenobacter hydrossis.
Biochemistry Mosc., 88, 2023
|
|
8AHR
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense in holo form with PLP | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
8AHQ
 
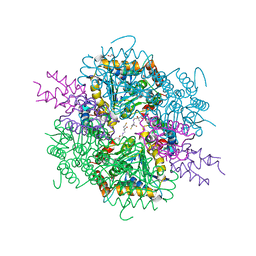 | | VirD/holo-ACP5b of Streptomyces virginiae complex | | Descriptor: | 1,2-ETHANEDIOL, 4'-PHOSPHOPANTETHEINE, CHLORIDE ION, ... | | Authors: | Collin, S, Gruez, A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8AHM
 
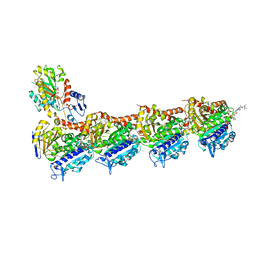 | | Crystal structure of tubulin in complex with C(13)/C(13')-Bis-Desmethyl-Disorazole Z | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Oliva, M.A, Diaz, J.F, Altmann, K.H. | | Deposit date: | 2022-07-22 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Synthesis and Biological Evaluation of C(13)/C(13')-Bis(desmethyl)disorazole Z.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8AHG
 
 | |
8AHD
 
 | |
8AHC
 
 | | Crystal structure of the BRD9 bromodomain with BI-7189 | | Descriptor: | Bromodomain-containing protein 9, [2,6-dimethoxy-4-(1,2,5-trimethyl-6-oxidanylidene-pyridin-3-yl)phenyl]methyl-dimethyl-azanium | | Authors: | Bader, G, Boettcher, J, Weiss-Puxbaum, A. | | Deposit date: | 2022-07-21 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Discovery of a Chemical Probe to Study Implications of BPTF Bromodomain Inhibition in Cellular and in vivo Experiments.
Chemmedchem, 18, 2023
|
|
8AHB
 
 | |
8AHA
 
 | | rsEGFP2 photoswitched to its off-state at 100K | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Mantovanelli, A, Adam, V. | | Deposit date: | 2022-07-21 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Photophysical Studies at Cryogenic Temperature Reveal a Novel Photoswitching Mechanism of rsEGFP2.
J.Am.Chem.Soc., 145, 2023
|
|
8AH3
 
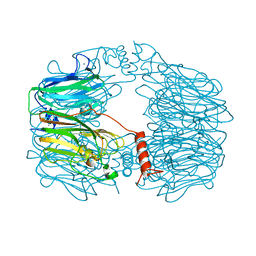 | |
8AH1
 
 | | BK Polyomavirus VP1 mutant N-Q | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1 | | Authors: | Sorin, M.N, Di Maio, A, Silva, L.M, Ebert, D, Delannoy, C, Nguyen, N.-K, Guerardel, Y, Chai, W, Halary, F, Renaudin-Autain, K, Liu, Y, Bressollette-Bodin, C, Stehle, T, McIlroy, D. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structural and functional analysis of natural capsid variants suggests sialic acid-independent entry of BK polyomavirus.
Cell Rep, 42, 2023
|
|
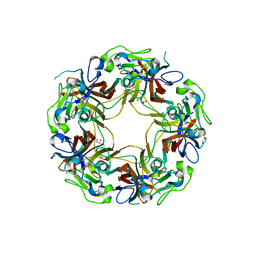
8AH0
 
 | | BK Polyomavirus VP1 mutant VQQ | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1 | | Authors: | Sorin, M.N, Di Maio, A, Silva, L.M, Ebert, D, Delannoy, C, Nguyen, N.-K, Guerardel, Y, Chai, W, Halary, F, Renaudin-Autain, K, Liu, Y, Bressollette-Bodin, C, Stehle, T, McIlroy, D. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural and functional analysis of natural capsid variants suggests sialic acid-independent entry of BK polyomavirus.
Cell Rep, 42, 2023
|
|
8AGY
 
 | |
8AGO
 
 | | BK Polyomavirus VP1 mutant E73Q | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1 | | Authors: | Sorin, M.N, Di Maio, A, Silva, L.M, Ebert, D, Delannoy, C, Nguyen, N.-K, Guerardel, Y, Chai, W, Halary, F, Renaudin-Autain, K, Liu, Y, Bressollette-Bodin, C, Stehle, T, McIlroy, D. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Structural and functional analysis of natural capsid variants suggests sialic acid-independent entry of BK polyomavirus.
Cell Rep, 42, 2023
|
|
8AGL
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R in complex with JMC31 | | Descriptor: | 1-[2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-5-[4-[(cyclohexylmethylamino)methyl]phenyl]-~{N}-ethyl-1,2,3-triazole-4-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mazzorana, M, Mangani, S, Maramai, S. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of Human Heat Shock Protein 90 N-Terminal Domain and Its Variants K112R and K112A in Complex with a Potent 1,2,3-Triazole-Based Inhibitor.
Int J Mol Sci, 23, 2022
|
|
8AGJ
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112A in complex with JMC31 | | Descriptor: | 1-[2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-5-[4-[(cyclohexylmethylamino)methyl]phenyl]-~{N}-ethyl-1,2,3-triazole-4-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mazzorana, M, Mangani, S, Maramai, S. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Characterization of Human Heat Shock Protein 90 N-Terminal Domain and Its Variants K112R and K112A in Complex with a Potent 1,2,3-Triazole-Based Inhibitor.
Int J Mol Sci, 23, 2022
|
|
8AGI
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) in complex with JMC31 | | Descriptor: | 1-[2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-5-[4-[(cyclohexylmethylamino)methyl]phenyl]-~{N}-ethyl-1,2,3-triazole-4-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mazzorana, M, Mangani, S, Maramai, S. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Human Heat Shock Protein 90 N-Terminal Domain and Its Variants K112R and K112A in Complex with a Potent 1,2,3-Triazole-Based Inhibitor.
Int J Mol Sci, 23, 2022
|
|
8AGH
 
 | | BK Polyomavirus VP1 mutant E73A | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1 | | Authors: | Sorin, M.N, Di Maio, A, Silva, L.M, Ebert, D, Delannoy, C, Nguyen, N.-K, Guerardel, Y, Chai, W, Halary, F, Renaudin-Autain, K, Liu, Y, Bressollette-Bodin, C, Stehle, T, McIlroy, D. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Structural and functional analysis of natural capsid variants suggests sialic acid-independent entry of BK polyomavirus.
Cell Rep, 42, 2023
|
|
8AGD
 
 | | Full SDBC and SOD assembly | | Descriptor: | (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,16,20,24-hexamethyl-24-oxidanyl-pentacosyl]-4,4,6-trimethyl-cyclohexane-1,3-diol, COPPER (II) ION, FE (III) ION, ... | | Authors: | Farci, D, Graca, A.T, Piano, D. | | Deposit date: | 2022-07-19 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The SDBC is active in quenching oxidative conditions and bridges the cell envelope layers in Deinococcus radiodurans.
J.Biol.Chem., 299, 2023
|
|
