4KB0
 
 | |
7P0V
 
 | | Crystal structure of human SF3A1 ubiquitin-like domain in complex with U1 snRNA stem-loop 4 | | Descriptor: | Isoform 2 of Splicing factor 3A subunit 1, PENTAETHYLENE GLYCOL, RNA (5'-R(P*GP*GP*GP*GP*AP*CP*UP*GP*CP*GP*UP*UP*CP*GP*CP*GP*CP*UP*UP*UP*CP*CP*CP*C)-3'), ... | | Authors: | Sabath, K, de Vries, T, Jonas, S. | | Deposit date: | 2021-06-30 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Sequence-specific RNA recognition by an RGG motif connects U1 and U2 snRNP for spliceosome assembly.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5ZTM
 
 | | Crystal structure of MLE dsRBDs in complex with roX2 (R2H1) | | Descriptor: | Dosage compensation regulator, non-coding mRNA sequence roX2 | | Authors: | Lv, M.Q, Tang, Y.J. | | Deposit date: | 2018-05-04 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural insights reveal the specific recognition of roX RNA by the dsRNA-binding domains of the RNA helicase MLE and its indispensable role in dosage compensation in Drosophila.
Nucleic Acids Res., 47, 2019
|
|
6EEC
 
 | | Mycobacterium tuberculosis RNAP promoter unwinding intermediate complex with RbpA/CarD and AP3 promoter captured by Corallopyronin | | Descriptor: | DNA (63-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
4WYL
 
 | | Mutant K18E of 3D polymerase from Foot-and-Moth Disease Virus | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Ferrer-Orta, C, Verdaguer, N, de la Higuera, I, Domingo, E. | | Deposit date: | 2014-11-17 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multifunctionality of a picornavirus polymerase domain: nuclear localization signal and nucleotide recognition.
J.Virol., 89, 2015
|
|
6EDT
 
 | | Mycobacterium tuberculosis RNAP open promoter complex with RbpA/CarD and AP3 promoter | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
6EE8
 
 | | Mycobacterium tuberculosis RNAP promoter unwinding intermediate complex with RbpA/CarD and AP3 promoter | | Descriptor: | DNA (60-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
8PM0
 
 | |
8CRO
 
 | | Cryo-EM structure of Pyrococcus furiosus transcription elongation complex | | Descriptor: | DNA Non-Template Strand, DNA Template Strand, DNA-directed RNA polymerase subunit Rpo10, ... | | Authors: | Tarau, D.M, Grunberger, F, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-03-08 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
8R3K
 
 | |
8I16
 
 | | Crystal structure of the selenomethionine (SeMet)-derived Cas12g (D513A) mutant | | Descriptor: | Cas12g, ZINC ION | | Authors: | Zhang, B, Chen, J, Ye, Y.M, OuYang, S.Y. | | Deposit date: | 2023-01-12 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural transitions upon guide RNA binding and their importance in Cas12g-mediated RNA cleavage.
Plos Genet., 19, 2023
|
|
8POH
 
 | |
7UBM
 
 | |
6JCY
 
 | |
7UBN
 
 | |
3JB9
 
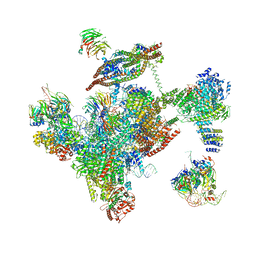 | | Cryo-EM structure of the yeast spliceosome at 3.6 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, C, Hang, J, Wan, R, Huang, M, Wong, C, Shi, Y. | | Deposit date: | 2015-08-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a yeast spliceosome at 3.6-angstrom resolution
Science, 349, 2015
|
|
8X6G
 
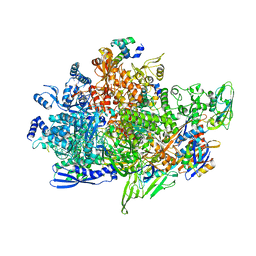 | | Cryo-EM structure of Staphylococcus aureus sigB-dependent RNAP-promoter open complex | | Descriptor: | DNA (70-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yuan, L, Xu, L, Liu, Q, Feng, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase.
Nat Commun, 15, 2024
|
|
8GZQ
 
 | | Cryo-EM structure of the NS5-NS3-SLA complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Genome polyprotein, RNA, ... | | Authors: | Osawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2022-09-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of dengue virus RNA replicase complexes.
Mol.Cell, 83, 2023
|
|
1PVO
 
 | |
8R3L
 
 | | Influenza A/H7N9 polymerase in pre-initiation state, intermediate conformation (I) with PB2-C(I), ENDO(T), and Pol II pS5 CTD peptide mimic bound in site 1A/2A | | Descriptor: | 3' vRNA end (51-mer vRNA loop), Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2023-11-09 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The host RNA polymerase II C-terminal domain is the anchor for replication of the influenza virus genome.
Nat Commun, 15, 2024
|
|
7VF9
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
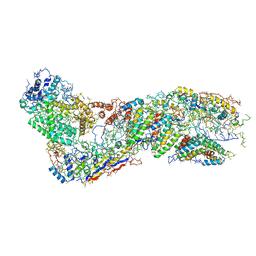
6O1O
 
 | |
7XL4
 
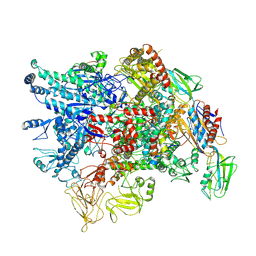 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (closed lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7XL3
 
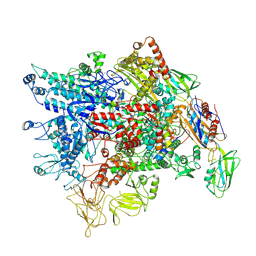 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (open lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
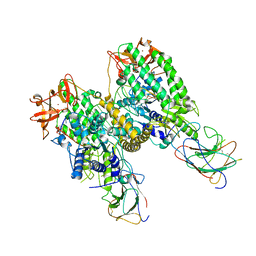
8G7U
 
 | |
