1HMV
 
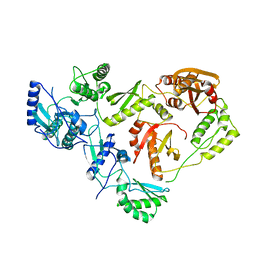 | | THE STRUCTURE OF UNLIGANDED REVERSE TRANSCRIPTASE FROM THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66), MAGNESIUM ION | | Authors: | Rodgers, D.W, Gamblin, S.J, Harris, B.A, Ray, S, Culp, J.S, Hellmig, B, Woolf, D.J, Debouck, C, Harrison, S.C. | | Deposit date: | 1994-12-15 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1P5N
 
 | |
2JZE
 
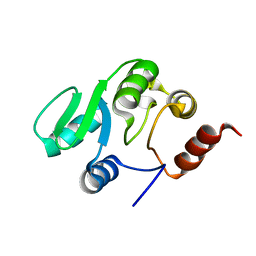 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3, single conformer closest to the mean coordinates of an ensemble of twenty energy minimized conformers | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
8T2T
 
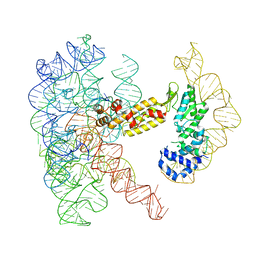 | | Structure of a group II intron ribonucleoprotein in the post-ligation (post-2F) state | | Descriptor: | AMMONIUM ION, Group II intron reverse transcriptase/maturase, MAGNESIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
6O3W
 
 | |
6WWA
 
 | | Crystal structure of human SHLD2-SHLD3-REV7 complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 2,Shieldin complex subunit 3 chimera | | Authors: | Xie, W, Patel, D.J. | | Deposit date: | 2020-05-08 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WW9
 
 | | Crystal structure of human REV7(R124A)-SHLD3(35-58) complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 3 | | Authors: | Xie, W, Patel, D.J. | | Deposit date: | 2020-05-08 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1YG3
 
 | |
2WFS
 
 | | Fitting of influenza virus NP structure into the 9-fold symmetryzed cryoEM reconstruction of an active RNP particle. | | Descriptor: | NUCLEOPROTEIN | | Authors: | Coloma, R, Valpuesta, J.M, Arranz, R, Carrascosa, J.L, Ortin, J, Martin-Benito, J. | | Deposit date: | 2009-04-15 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | The Structure of a Biologically Active Influenza Virus Ribonucleoprotein Complex.
Plos Pathog., 5, 2009
|
|
3ODR
 
 | |
2JZD
 
 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
5OP0
 
 | |
4QPL
 
 | | Crystal structure of RNF146(RING-WWE)/UbcH5a/iso-ADPr complex | | Descriptor: | 2'-O-(5-O-phosphono-alpha-D-ribofuranosyl)adenosine 5'-(dihydrogen phosphate), E3 ubiquitin-protein ligase RNF146, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Wang, Z, DaRosa, P.A, Klevit, R.E, Xu, W. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal.
Nature, 517, 2015
|
|
4XW6
 
 | | X-ray structure of PKAc with ADP, free phosphate ion, CP20, magnesium ions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | Deposit date: | 2015-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
4XW5
 
 | | X-ray structure of PKAc with ATP, CP20, calcium ions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | Deposit date: | 2015-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
6IKA
 
 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:entecavir-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6IK9
 
 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
1WBO
 
 | | fragment based p38 inhibitors | | Descriptor: | 2-CHLOROPHENOL, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Cleasby, A, Devine, L, Gill, A, Jhoti, H. | | Deposit date: | 2004-11-04 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
4XW4
 
 | | X-ray structure of PKAc with AMPPNP, SP20, calcium ions | | Descriptor: | CALCIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | Deposit date: | 2015-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2015-07-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
1W7H
 
 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | 3-(BENZYLOXY)PYRIDIN-2-AMINE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Jhoti, H, Gill, A, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-02 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
8T2S
 
 | | Structure of a group II intron ribonucleoprotein in the pre-branching (pre-1F) state | | Descriptor: | AMMONIUM ION, CALCIUM ION, Group II intron reverse transcriptase/maturase, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
6R3V
 
 | | Crystal Structure of RhoA-GDP-Pi in Complex with RhoGAP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jin, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A GAP-GTPase-GDP-PiIntermediate Crystal Structure Analyzed by DFT Shows GTP Hydrolysis Involves Serial Proton Transfers.
Chemistry, 25, 2019
|
|
6KDM
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:entecavir 5'-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDN
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDJ
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:lamivudine 5'-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
