4J9H
 
 | |
1HAQ
 
 | |
1GWD
 
 | | Tri-iodide derivative of hen egg-white lysozyme | | Descriptor: | 1,2-ETHANEDIOL, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Evans, G, Bricogne, G. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-06 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Triiodide Derivatization and Combinatorial Counter-Ion Replacement: Two Methods for Enhancing Phasing Signal Using Laboratory Cu Kalpha X-Ray Equipment
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4J9C
 
 | |
3GTE
 
 | | Crystal Structure of Dicamba Monooxygenase with Non-heme Iron | | Descriptor: | ACETATE ION, DdmC, FE (III) ION, ... | | Authors: | Rydel, T.J, Sturman, E.J, Moshiri, F, Brown, G.R, Qi, Y. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dicamba monooxygenase: structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation.
J.Mol.Biol., 392, 2009
|
|
1LPB
 
 | | THE 2.46 ANGSTROMS RESOLUTION STRUCTURE OF THE PANCREATIC LIPASE COLIPASE COMPLEX INHIBITED BY A C11 ALKYL PHOSPHONATE | | Descriptor: | CALCIUM ION, COLIPASE, LIPASE, ... | | Authors: | Egloff, M.-P, Van Tilbeurgh, H, Cambillau, C. | | Deposit date: | 1994-08-19 | | Release date: | 1994-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The 2.46 A resolution structure of the pancreatic lipase-colipase complex inhibited by a C11 alkyl phosphonate.
Biochemistry, 34, 1995
|
|
4J9G
 
 | |
1LPM
 
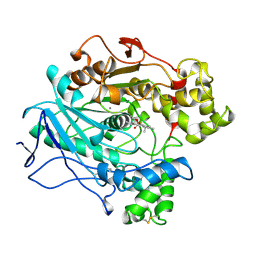 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | Descriptor: | (1R)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grochulski, P.G, Cygler, M.C. | | Deposit date: | 1995-01-06 | | Release date: | 1995-04-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
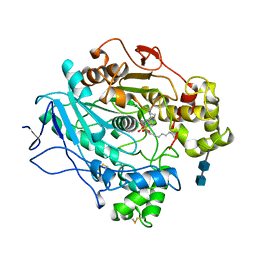
1LPN
 
 | |
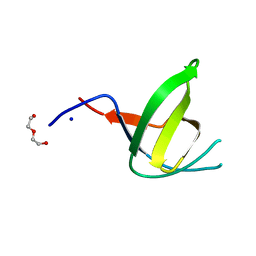
4JJD
 
 | |
2QFD
 
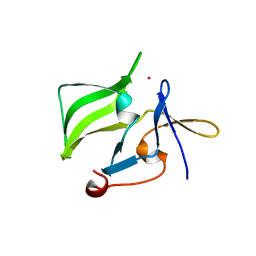 | | Crystal structure of the regulatory domain of human RIG-I with bound Hg | | Descriptor: | MERCURY (II) ION, Probable ATP-dependent RNA helicase DDX58 | | Authors: | Cui, S, Lammens, A, Lammens, K, Hopfner, K.P. | | Deposit date: | 2007-06-27 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The C-Terminal Regulatory Domain Is the RNA 5'-Triphosphate Sensor of RIG-I.
Mol.Cell, 29, 2008
|
|
1VLK
 
 | |
1ITV
 
 | | Dimeric form of the haemopexin domain of MMP9 | | Descriptor: | MMP9, SULFATE ION | | Authors: | Cha, H, Kopetzki, E, Huber, R, Lanzendoerfer, M, Brandstetter, H. | | Deposit date: | 2002-02-11 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of the adaptive molecular recognition by MMP9.
J.Mol.Biol., 320, 2002
|
|
3GTS
 
 | | Crystal Structure of Dicamba Monooxygenase with Non-heme Iron and Dicamba | | Descriptor: | 3,6-dichloro-2-methoxybenzoic acid, DdmC, FE (III) ION, ... | | Authors: | Rydel, T.J, Sturman, E.J, Moshiri, F, Brown, G.R, Qi, Y. | | Deposit date: | 2009-03-28 | | Release date: | 2009-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dicamba monooxygenase: structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation.
J.Mol.Biol., 392, 2009
|
|
1I16
 
 | | STRUCTURE OF INTERLEUKIN 16: IMPLICATIONS FOR FUNCTION, NMR, 20 STRUCTURES | | Descriptor: | INTERLEUKIN 16 | | Authors: | Muehlhahn, P, Zweckstetter, M, Georgescu, J, Ciosto, C, Renner, C, Lanzendoerfer, M, Lang, K, Ambrosius, D, Baier, M, Kurth, R, Holak, T.A. | | Deposit date: | 1998-05-20 | | Release date: | 1999-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of interleukin 16 resembles a PDZ domain with an occluded peptide binding site.
Nat.Struct.Biol., 5, 1998
|
|
3GOB
 
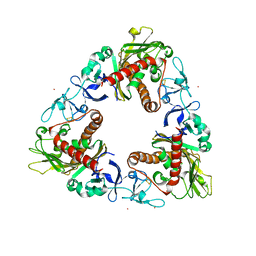 | | Crystal Structure of Dicamba Monooxygenase with Non-heme Cobalt and DCSA | | Descriptor: | 3,6-dichloro-2-hydroxybenzoic acid, COBALT (II) ION, DdmC, ... | | Authors: | Rydel, T.J, Sturman, E.J, Moshiri, F, Brown, G.R, Qi, Y. | | Deposit date: | 2009-03-18 | | Release date: | 2009-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dicamba monooxygenase: structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation.
J.Mol.Biol., 392, 2009
|
|
4JNW
 
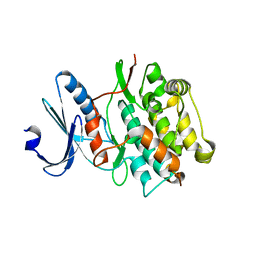 | | Bacterially expressed Titin Kinase | | Descriptor: | GLYCEROL, Titin | | Authors: | Bogomolovas, J, Labeit, S, Mayans, O. | | Deposit date: | 2013-03-16 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Titin kinase is an inactive pseudokinase scaffold that supports MuRF1 recruitment to the sarcomeric M-line.
Open Biol, 4, 2014
|
|
4JJC
 
 | |
4KUK
 
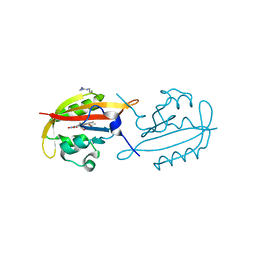 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Dark state) | | Descriptor: | ACETIC ACID, RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
4LQU
 
 | | 1.60A resolution crystal structure of a superfolder green fluorescent protein (W57G) mutant | | Descriptor: | Green fluorescent protein | | Authors: | Lovell, S, Xia, Y, Vo, B, Battaile, K.P, Egan, C, Karanicolas, J. | | Deposit date: | 2013-07-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The designability of protein switches by chemical rescue of structure: mechanisms of inactivation and reactivation.
J.Am.Chem.Soc., 135, 2013
|
|
4LCR
 
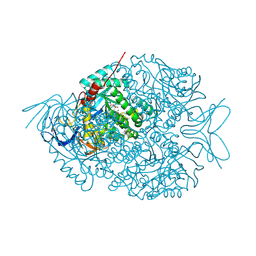 | | The crystal structure of di-Zn dihydropyrimidinase in complex with NCBA | | Descriptor: | Chromosome 8 SCAF14545, whole genome shotgun sequence, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2013-06-22 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
2J62
 
 | | Structure of a bacterial O-glcnacase in complex with glcnacstatin | | Descriptor: | CHLORIDE ION, N-[(5R,6R,7R,8S)-6,7-DIHYDROXY-5-(HYDROXYMETHYL)-2-(2-PHENYLETHYL)-1,5,6,7,8,8A-HEXAHYDROIMIDAZO[1,2-A]PYRIDIN-8-YL]-2-METHYLPROPANAMIDE, O-GlcNAcase NagJ | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Schimpl, M, Shepherd, S.M, Shpiro, N.A, van Aalten, D.M.F. | | Deposit date: | 2006-09-22 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels.
J. Am. Chem. Soc., 128, 2006
|
|
4LCS
 
 | | The crystal structure of di-Zn dihydropyrimidinase in complex with hydantoin | | Descriptor: | Chromosome 8 SCAF14545, whole genome shotgun sequence, ZINC ION, ... | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S. | | Deposit date: | 2013-06-23 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
4MMG
 
 | | crystal structure of YafQ mutant H87Q from E.coli | | Descriptor: | SULFATE ION, mRNA interferase YafQ | | Authors: | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2013-09-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
4ML2
 
 | | Crystal structure of wild-type YafQ | | Descriptor: | SULFATE ION, mRNA interferase YafQ | | Authors: | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2013-09-06 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
