8DVY
 
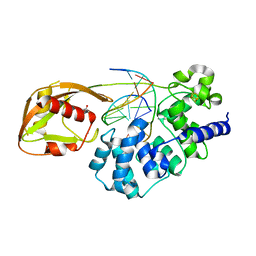 | | DNA glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with an enzyme-generated abasic site product (AP) and crystalized with calcium acetate | | Descriptor: | ACETATE ION, Adenine DNA glycosylase, CALCIUM ION, ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-30 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
6UEU
 
 | |
6E49
 
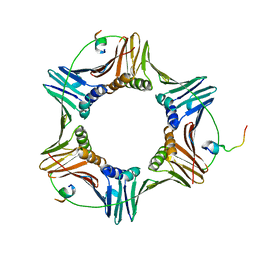 | | Pif1 peptide bound to PCNA trimer | | Descriptor: | ATP-dependent DNA helicase PIF1, Proliferating cell nuclear antigen | | Authors: | Buzovetsky, O, Kwon, Y, Pham, N.T, Kim, C, Ira, G, Sung, P, Xiong, Y. | | Deposit date: | 2018-07-17 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of the Pif1-PCNA Complex in Pol delta-Dependent Strand Displacement DNA Synthesis and Break-Induced Replication.
Cell Rep, 21, 2017
|
|
9FOY
 
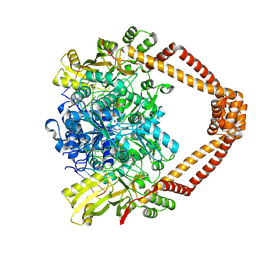 | | Ternary complex of a Mycobacterium tuberculosis DNA gyrase core fusion with DNA and the inhibitor AMK32b | | Descriptor: | (1~{R})-2-[4-[[4-bromanyl-3,5-bis(fluoranyl)phenyl]methylamino]cyclohexyl]-1-(6-methoxy-1,5-naphthyridin-4-yl)ethanol, (1~{S})-2-[4-[[4-bromanyl-3,5-bis(fluoranyl)phenyl]methylamino]cyclohexyl]-1-(6-methoxy-1,5-naphthyridin-4-yl)ethanol, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G)-3'), ... | | Authors: | Kokot, M, Hrast, M, Feng, L, Mitchenall, L.A, Lawson, D.M, Maxwell, A, Minovski, N, Anderluh, M. | | Deposit date: | 2024-06-12 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unraveling the Mycobacterium tuberculosis DNA gyrase-DNA-NBTI complex
To be published
|
|
3SGI
 
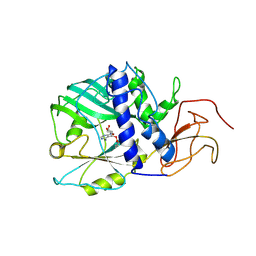 | |
7OPD
 
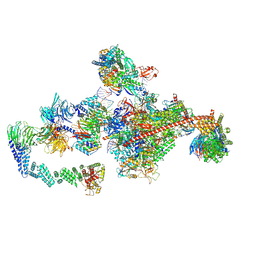 | | Pol II-CSB-CRL4CSA-UVSSA-SPT6-PAF (Structure 5) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-31 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OPC
 
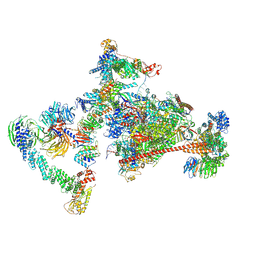 | | Pol II-CSB-CRL4CSA-UVSSA-SPT6-PAF (Structure 4) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-31 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
3SC3
 
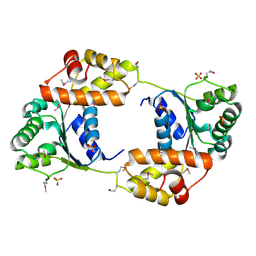 | |
9GDM
 
 | |
9GGQ
 
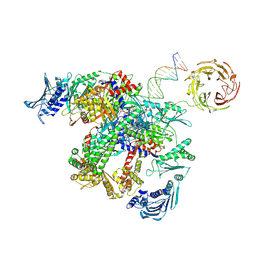 | | E.coli gyrase holocomplex with cleaved chirally wrapped 217 bp DNA fragment and moxifloxacin | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Ghilarov, D, Heddle, J.G, Pabis, M. | | Deposit date: | 2024-08-13 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of chiral wrap and T-segment capture by Escherichia coli DNA gyrase
Proceedings of the National Academy of Sciences USA, 2024
|
|
7OO3
 
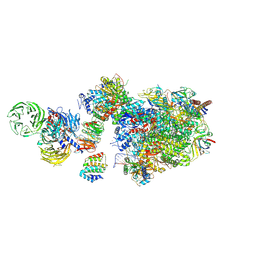 | | Pol II-CSB-CSA-DDB1-UVSSA (Structure1) | | Descriptor: | CSB element, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-26 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OOB
 
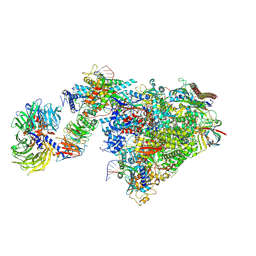 | | Pol II-CSB-CSA-DDB1-UVSSA-ADPBeF3 (Structure2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA damage-binding protein 1, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
6U8Q
 
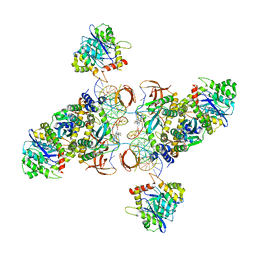 | | CryoEM structure of HIV-1 cleaved synaptic complex (CSC) intasome | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Li, M, Chen, X, Craigie, R. | | Deposit date: | 2019-09-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | A Peptide Derived from Lens Epithelium-Derived Growth Factor Stimulates HIV-1 DNA Integration and Facilitates Intasome Structural Studies.
J.Mol.Biol., 432, 2020
|
|
7M9A
 
 | | ADP-AlF3 bound TnsC structure from ShCAST system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (27-MER), TnsC | | Authors: | Park, J, Tsai, A.W.L, Mehrotra, E, Kellogg, E.H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for target site selection in RNA-guided DNA transposition systems.
Science, 373, 2021
|
|
7M9B
 
 | | ADP-AlF3 bound TnsC structure in closed form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (27-MER), TnsC | | Authors: | Park, J, Tsai, A.W.L, Mehrotra, E, Kellogg, E.H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for target site selection in RNA-guided DNA transposition systems.
Science, 373, 2021
|
|
5CP6
 
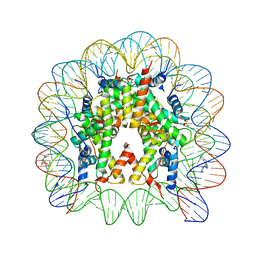 | | Nucleosome Core Particle with Adducts from the Anticancer Compound, [(eta6-5,8,9,10-tetrahydroanthracene)Ru(ethylenediamine)Cl][PF6] | | Descriptor: | (ethane6-5,8,9,10-tetrahydroanthracene)Ru(II)(ethylene-diamine)Cl, DNA (145-MER), Histone H2A, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2015-07-21 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An Organometallic Compound which Exhibits a DNA Topology-Dependent One-Stranded Intercalation Mode.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6OPM
 
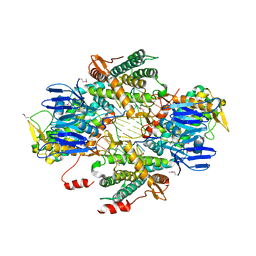 | | Casposase bound to integration product | | Descriptor: | CALCIUM ION, CRISPR-associated endonuclease Cas1, DNA 21-mer, ... | | Authors: | Dyda, F, Hickman, A.B, Kailasan, S. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-12 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Casposase structure and the mechanistic link between DNA transposition and spacer acquisition by CRISPR-Cas.
Elife, 9, 2020
|
|
2EE1
 
 | | Solution structures of the Chromo domain of human chromodomain helicase-DNA-binding protein 4 | | Descriptor: | Chromodomain helicase-DNA-binding protein 4 | | Authors: | Sato, M, Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-15 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the Chromo domain of human chromodomain helicase-DNA-binding protein 4
To be Published
|
|
4WXD
 
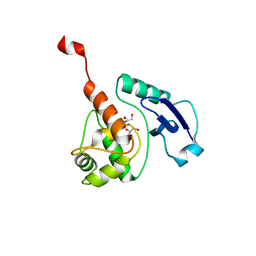 | | Crystal structure of Mycobacterium tuberculosis OGT-R37K | | Descriptor: | GLYCEROL, Methylated-DNA--protein-cysteine methyltransferase | | Authors: | Miggiano, R, Rossi, F, Garavaglia, S, Rizzi, M. | | Deposit date: | 2014-11-13 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis O6-methylguanine-DNA methyltransferase protein clusters assembled on to damaged DNA.
Biochem.J., 473, 2016
|
|
4WXC
 
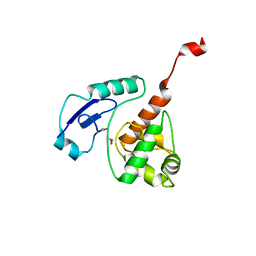 | | Crystal structure of Mycobacterium tuberculosis OGT-Y139F | | Descriptor: | 1,2-ETHANEDIOL, Methylated-DNA--protein-cysteine methyltransferase | | Authors: | Miggiano, R, Rossi, F, Garavaglia, S, Rizzi, M. | | Deposit date: | 2014-11-13 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis O6-methylguanine-DNA methyltransferase protein clusters assembled on to damaged DNA.
Biochem.J., 473, 2016
|
|
6EO3
 
 | |
6EO2
 
 | |
9B8U
 
 | |
7MK1
 
 | | Structure of a protein-modified aptamer complex | | Descriptor: | Antiviral innate immune response receptor RIG-I, DNA (41-MER), MAGNESIUM ION, ... | | Authors: | Ren, X, Pyle, A.M. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolving A RIG-I Antagonist: A Modified DNA Aptamer Mimics Viral RNA.
J.Mol.Biol., 433, 2021
|
|
7O4I
 
 | | Yeast RNA polymerase II transcription pre-initiation complex with initial transcription bubble | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | Deposit date: | 2021-04-06 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
