6IIW
 
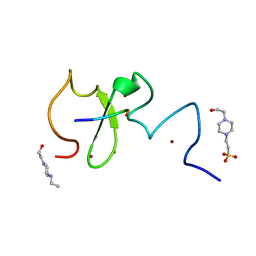 | | Crystal structure of human UHRF1 PHD finger in complex with PAF15 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase UHRF1, PCNA-associated factor, ... | | Authors: | Arita, K, Kori, S. | | Deposit date: | 2018-10-07 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Two distinct modes of DNMT1 recruitment ensure stable maintenance DNA methylation.
Nat Commun, 11, 2020
|
|
8HSV
 
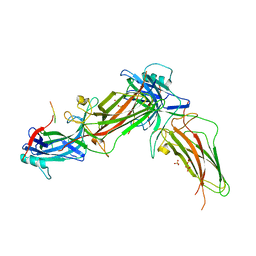 | | The structure of rat beta-arrestin1 in complex with a rat Mdm2 peptide | | Descriptor: | Beta-arrestin-1, SULFATE ION, peptide from E3 ubiquitin-protein ligase Mdm2 | | Authors: | Yun, Y, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GPCR targeting of E3 ubiquitin ligase MDM2 by inactive beta-arrestin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6NVO
 
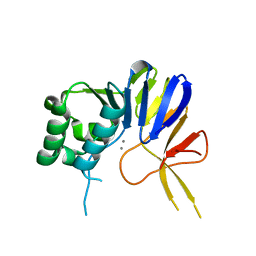 | | Crystal structure of Pseudomonas putida nuclease MPE | | Descriptor: | MANGANESE (II) ION, Nuclease MPE | | Authors: | Goldgur, Y, Shuman, S, Ejaz, A. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Activity and structure ofPseudomonas putidaMPE, a manganese-dependent single-strand DNA endonuclease encoded in a nucleic acid repair gene cluster.
J.Biol.Chem., 294, 2019
|
|
6NVP
 
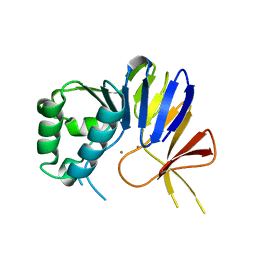 | | Crystal structure of Pseudomonas putida nuclease MPE | | Descriptor: | MANGANESE (II) ION, Nuclease MPE | | Authors: | Goldgur, Y, Shuman, S, Ejaz, A. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity and structure ofPseudomonas putidaMPE, a manganese-dependent single-strand DNA endonuclease encoded in a nucleic acid repair gene cluster.
J.Biol.Chem., 294, 2019
|
|
5ZE2
 
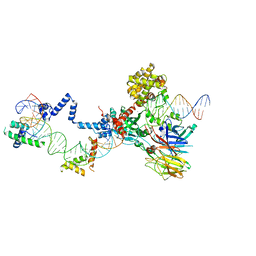 | | Hairpin Complex, RAG1/2-hairpin 12RSS/23RSS complex in 5mM Mn2+ for 2 min at 4'C | | Descriptor: | 1,2-ETHANEDIOL, DNA (30-MER), DNA (31-MER), ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
2YBF
 
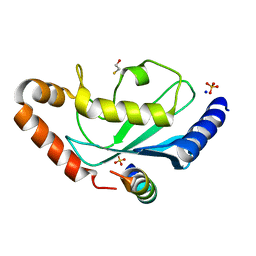 | | Complex of Rad18 (Rad6 binding domain) with Rad6b | | Descriptor: | BETA-MERCAPTOETHANOL, E3 UBIQUITIN-PROTEIN LIGASE RAD18, SODIUM ION, ... | | Authors: | Hibbert, R.G, Sixma, T.K. | | Deposit date: | 2011-03-08 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | E3 Ligase Rad18 Promotes Monoubiquitination Rather Than Ubiquitin Chain Formation by E2 Enzyme Rad6.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5NWL
 
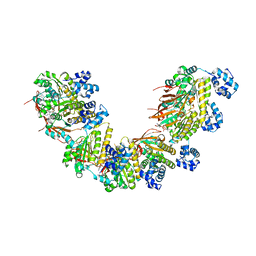 | | Crystal structure of a human RAD51-ATP filament. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA repair protein RAD51 homolog 1, MAGNESIUM ION | | Authors: | Pellegrini, L, Moschetti, T. | | Deposit date: | 2017-05-06 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Two distinct conformational states define the interaction of human RAD51-ATP with single-stranded DNA.
EMBO J., 37, 2018
|
|
6CIJ
 
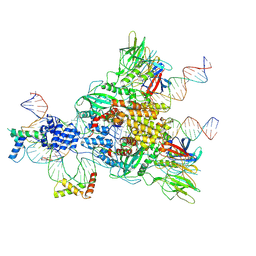 | | Cryo-EM structure of mouse RAG1/2 HFC complex containing partial HMGB1 linker(3.9 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
3RD2
 
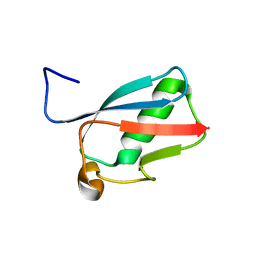 | | NIP45 SUMO-like Domain 2 | | Descriptor: | NFATC2-interacting protein | | Authors: | Perry, J.J.P, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes.
Mol.Cell.Biol., 31, 2011
|
|
5ZE1
 
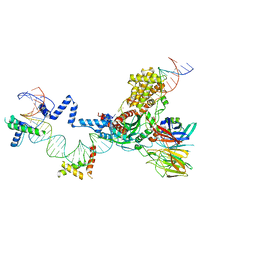 | | Hairpin Forming Complex, RAG1/2-Nicked 12RSS/23RSS complex in 2mM Mn2+ for 10 min at 4'C | | Descriptor: | 1,2-ETHANEDIOL, DNA, HMGB1 A-B box, ... | | Authors: | Kim, M.S, Chuenchor, W, Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2018-02-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination
Mol. Cell, 70, 2018
|
|
6CG0
 
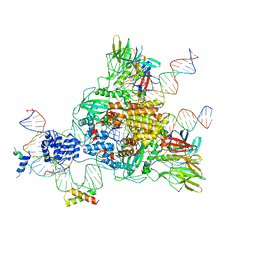 | | Cryo-EM structure of mouse RAG1/2 HFC complex (3.17 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
3RKX
 
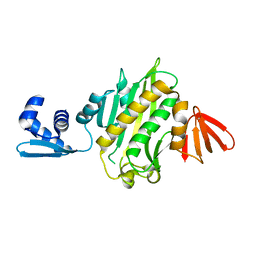 | |
3RKW
 
 | |
8G7U
 
 | |
8G7T
 
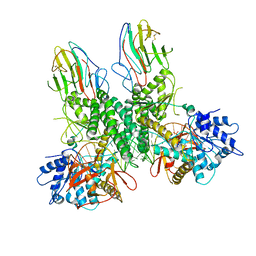 | | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-end) | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The E3 ligase Riplet promotes RIG-I signaling independent of RIG-I oligomerization.
Nat Commun, 14, 2023
|
|
8G7V
 
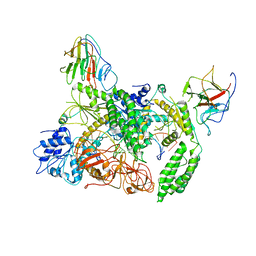 | | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-inter) | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The E3 ligase Riplet promotes RIG-I signaling independent of RIG-I oligomerization.
Nat Commun, 14, 2023
|
|
3FL2
 
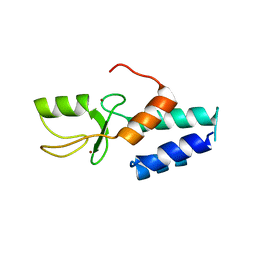 | | Crystal structure of the ring domain of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ring Domain of the E3 Ubiquitin-Protein Ligase Uhrf1
To be Published
|
|
2K7F
 
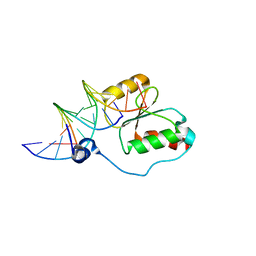 | | HADDOCK calculated model of the complex between the BRCT region of RFC p140 and dsDNA | | Descriptor: | 5'-D(P*DCP*DGP*DAP*DCP*DCP*DTP*DCP*DGP*DAP*DGP*DAP*DTP*DCP*DA)-3', 5'-D(P*DCP*DTP*DCP*DGP*DAP*DGP*DGP*DTP*DCP*DG)-3', Replication factor C subunit 1 | | Authors: | Kobayashi, M, Ab, E, Bonvin, A, Siegal, G. | | Deposit date: | 2008-08-10 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the DNA-bound BRCA1 C-terminal region from human replication factor C p140 and model of the protein-DNA complex.
J.Biol.Chem., 285, 2010
|
|
2MRE
 
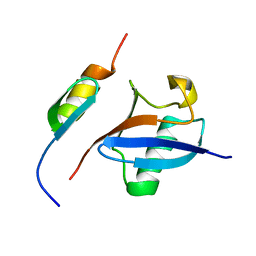 | | NMR structure of the Rad18-UBZ/ubiquitin complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Polyubiquitin-C, ZINC ION | | Authors: | Rizzo, A.A, Salerno, P.E, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response.
Biochemistry, 53, 2014
|
|
3GNA
 
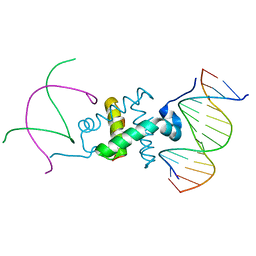 | | Crystal structure of the RAG1 nonamer-binding domain with DNA | | Descriptor: | 5'-D(*AP*CP*TP*TP*AP*AP*CP*AP*AP*AP*AP*AP*CP*C)-3', 5'-D(*TP*GP*GP*TP*TP*TP*TP*TP*GP*TP*TP*AP*AP*G)-3', V(D)J recombination-activating protein 1 | | Authors: | Yin, F.F, Bailey, S, Innis, C.A, Steitz, T.A, Schatz, D.G. | | Deposit date: | 2009-03-16 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3GNB
 
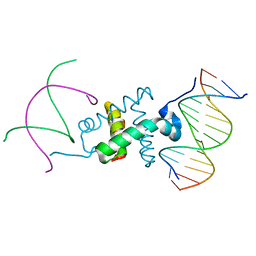 | | Crystal structure of the RAG1 nonamer-binding domain with DNA | | Descriptor: | 5'-D(*AP*AP*TP*TP*TP*TP*CP*AP*GP*AP*AP*AP*CP*C)-3', 5'-D(*AP*GP*GP*TP*TP*TP*CP*TP*GP*AP*AP*AP*AP*C)-3', V(D)J recombination-activating protein 1 | | Authors: | Yin, F.F, Bailey, S, Innis, C.A, Steitz, T.A, Schatz, D.G. | | Deposit date: | 2009-03-16 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6BRQ
 
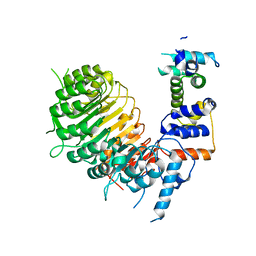 | | Crystal structure of rice ASK1-D3 ubiquitin ligase complex crystal form 3 | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-11-30 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
6BRO
 
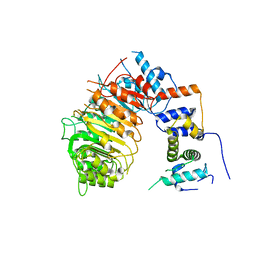 | |
4K95
 
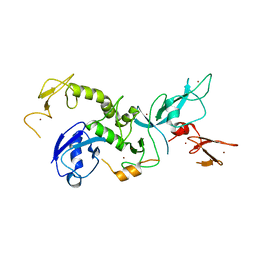 | | Crystal Structure of Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Seirafi, M, Menade, M, Sauve, V, Kozlov, G, Trempe, J.-F, Nagar, B, Gehring, K. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.499 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
7SA1
 
 | | LRR-F-Box plant ubiquitin ligase | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, F-box/LRR-repeat MAX2 homolog, ... | | Authors: | Palayam, M, Shabek, N. | | Deposit date: | 2021-09-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A conformational switch in the SCF-D3/MAX2 ubiquitin ligase facilitates strigolactone signalling.
Nat.Plants, 8, 2022
|
|
