2H2J
 
 | | Structure of Rubisco LSMT bound to Sinefungin and Monomethyllysine | | Descriptor: | N-METHYL-LYSINE, Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, SINEFUNGIN | | Authors: | Couture, J.F, Hauk, G, Trievel, R.C. | | Deposit date: | 2006-05-18 | | Release date: | 2006-05-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Catalytic Roles for Carbon-Oxygen Hydrogen Bonding in SET Domain Lysine Methyltransferases.
J.Biol.Chem., 281, 2006
|
|
5HXM
 
 | | Cycloalternan-forming enzyme from Listeria monocytogenes in complex with panose | | Descriptor: | Alpha-xylosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Winsor, J, Grimshaw, S, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-31 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5RBP
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library B03a | | Descriptor: | 2-[(1~{S})-1-azanylpropyl]phenol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
1XH6
 
 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | N-(4-{[4-(2-HYDROXY-5-PIPERIDIN-1-YLBENZOYL)BENZOYL]AMINO}AZEPAN-3-YL)ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
4FTG
 
 | |
2FXA
 
 | | Structure of the Protease Production Regulatory Protein hpr from Bacillus subtilis. | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-03 | | Release date: | 2006-03-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Protease Production Regulatory Protein hpr from Bacillus subtilis.
TO BE PUBLISHED
|
|
4FV5
 
 | | Crystal Structure of the ERK2 complexed with EK9 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 1, N-[(1S)-2-hydroxy-1-phenylethyl]-4-(5-methyl-2-phenylpyrimidin-4-yl)-1H-pyrrole-2-carboxamide, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Xie, X. | | Deposit date: | 2012-06-29 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the ERK2 complexed with EK9
TO BE PUBLISHED
|
|
4TV9
 
 | | Tubulin-PM060184 complex | | Descriptor: | (1Z,4S,6Z)-1-[(N-{(2Z,4Z,6E,8S)-8-[(2S)-5-methoxy-6-oxo-3,6-dihydro-2H-pyran-2-yl]-6-methylnona-2,4,6-trienoyl}-3-methy l-L-valyl)amino]octa-1,6-dien-4-yl carbamate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Bargsten, K, Diaz, J.F, Marsh, M, Cuevas, C, Liniger, M, Neuhaus, C, Andreu, J.M, Altmann, K.H, Steinmetz, M.O. | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1XM6
 
 | | Catalytic Domain Of Human Phosphodiesterase 4B In Complex With (R)-Mesopram | | Descriptor: | (5R)-5-(4-methoxy-3-propoxyphenyl)-5-methyl-1,3-oxazolidin-2-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-01 | | Release date: | 2004-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
4IVT
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, N-{N-[4-(acetylamino)-3,5-dichlorobenzyl]carbamimidoyl}-2-(1H-indol-1-yl)acetamide, SULFATE ION | | Authors: | Chen, T.T, Li, L, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2013-01-23 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Virtual screening and structure-based discovery of indole acylguanidines as potent beta-secretase (BACE1) inhibitors
Molecules, 18, 2013
|
|
1QNQ
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2,2':6',2''-TERPYRIDINE PLATINUM(II) Chloride, 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QSF
 
 | | STRUCTURE OF A6-TCR BOUND TO HLA-A2 COMPLEXED WITH ALTERED HTLV-1 TAX PEPTIDE Y8A | | Descriptor: | BETA-2 MICROGLOBULIN, HUMAN T-CELL RECEPTOR, MHC CLASS I HLA-A, ... | | Authors: | Ding, Y.H, Baker, B.M, Garboczi, D.N, Biddison, W.E, Wiley, D.C. | | Deposit date: | 1999-06-21 | | Release date: | 1999-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical.
Immunity, 11, 1999
|
|
3NY8
 
 | | Crystal structure of the human beta2 adrenergic receptor in complex with the inverse agonist ICI 118,551 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3S)-1-[(7-methyl-2,3-dihydro-1H-inden-4-yl)oxy]-3-[(1-methylethyl)amino]butan-2-ol, Beta-2 adrenergic receptor, ... | | Authors: | Wacker, D, Fenalti, G, Brown, M.A, Katritch, V, Abagyan, R, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Conserved Binding Mode of Human beta(2) Adrenergic Receptor Inverse Agonists and Antagonist Revealed by X-ray Crystallography
J.Am.Chem.Soc., 132, 2010
|
|
2OZF
 
 | | The crystal structure of the 2nd PDZ domain of the human NHERF-1 (SLC9A3R1) | | Descriptor: | Ezrin-radixin-moesin-binding phosphoprotein 50 | | Authors: | Phillips, C, Papagrigoriou, E, Gileadi, C, Fedorov, O, Elkins, J, Berridge, G, Turnbull, A.P, Gileadi, O, Schoch, G, Smee, C, Bray, J, Savitsky, P, Uppenberg, J, von Delft, F, Gorrec, F, Umeano, C, Salah, E, Colebrook, S, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the 2nd PDZ domain of the human NHERF-1 (SLC9A3R1)
To be Published
|
|
7D80
 
 | | Molecular model of the cryo-EM structure of 70S ribosome in complex with peptide deformylase, trigger factor, and methionine aminopeptidase | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Akbar, S, Bhakta, S, Sengupta, J. | | Deposit date: | 2020-10-06 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the interplay of protein biogenesis factors with the 70S ribosome.
Structure, 29, 2021
|
|
2ZWS
 
 | | Crystal Structure Analysis of neutral ceramidase from Pseudomonas aeruginosa | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kakuta, Y, Okino, N, Inoue, T, Okano, H, Ito, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase.
J.Biol.Chem., 284, 2009
|
|
4KNB
 
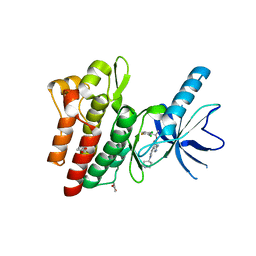 | | C-Met in complex with OSI ligand | | Descriptor: | 7-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[3,2-c]pyridin-6-amine, GAMMA-BUTYROLACTONE, Hepatocyte growth factor receptor | | Authors: | Wang, J, Steinig, A.G, Li, A.H, Chen, X, Dong, H, Ferraro, C, Jin, M, Kadalbajoo, M, Kleinberg, A, Stolz, K.M, Tavares-Greco, P.A, Wang, T, Albertella, M.R, Peng, Y, Crew, L, Kahler, J. | | Deposit date: | 2013-05-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel 6-aminofuro[3,2-c]pyridines as potent, orally efficacious inhibitors of cMET and RON kinases.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2ZVT
 
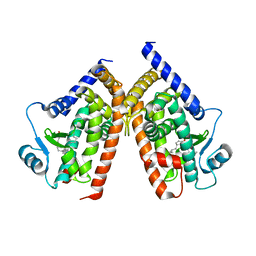 | | Cys285Ser mutant PPARgamma ligand-binding domain complexed with 15-deoxy-delta12,14-prostaglandin J2 | | Descriptor: | (5E,14E)-11-oxoprosta-5,9,12,14-tetraen-1-oic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Waku, T, Oyama, T, Shiraki, T, Morikawa, K. | | Deposit date: | 2008-11-19 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic structure of mutant PPARgamma LBD complexed with 15d-PGJ2: novel modulation mechanism of PPARgamma/RXRalpha function by covalently bound ligands
Febs Lett., 583, 2009
|
|
1XL9
 
 | | Crystal Structure of Dihydrodipicolinate Synthase DapA-2 (BA3935) from Bacillus Anthracis. | | Descriptor: | dihydrodipicolinate synthase | | Authors: | Blagova, E, Levdikov, V, Milioti, N, Fogg, M.J, Kalliomaa, A.K, Brannigan, J.A, Wilson, K.S, Wilkinson, A.J. | | Deposit date: | 2004-09-30 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of dihydrodipicolinate synthase (BA3935) from Bacillus anthracis at 1.94 A resolution.
Proteins, 62, 2006
|
|
2OTV
 
 | | Crystal structure of the complex formed between bovine trypsin and nicotinamide at 1.56 A resolution | | Descriptor: | CALCIUM ION, Cationic trypsin, NICOTINAMIDE, ... | | Authors: | Sinha, M, Singh, N, Sharma, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-02-09 | | Release date: | 2007-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the complex formed between bovine trypsin and nicotinamide at 1.56 A resolution
To be Published
|
|
2FYQ
 
 | |
6C65
 
 | | Crystal Structure of the Mango-II-A22U Fluorescent Aptamer Bound to TO1-Biotin | | Descriptor: | 4-[(3-{2-[(2-methoxyethyl)amino]-2-oxoethyl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, POTASSIUM ION, RNA (35-MER), ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-01-17 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.80005169 Å) | | Cite: | Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness.
Biochemistry, 57, 2018
|
|
4K05
 
 | |
1P7B
 
 | | Crystal structure of an inward rectifier potassium channel | | Descriptor: | POTASSIUM ION, integral membrane channel and cytosolic domains | | Authors: | Kuo, A, Gulbis, J.M, Antcliff, J.F, Rahman, T, Lowe, E.D, Zimmer, J, Cuthbertson, J, Ashcroft, F.M, Ezaki, T, Doyle, D.A. | | Deposit date: | 2003-05-01 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Crystal structure of the potassium channel KirBac1.1 in the closed state.
Science, 300, 2003
|
|
4OTF
 
 | | Crystal structure of the kinase domain of Bruton's Tyrosine kinase with GDC0834 | | Descriptor: | N-{3-[6-({4-[(2R)-1,4-dimethyl-3-oxopiperazin-2-yl]phenyl}amino)-4-methyl-5-oxo-4,5-dihydropyrazin-2-yl]-2-methylphenyl }-4,5,6,7-tetrahydro-1-benzothiophene-2-carboxamide, SULFATE ION, Tyrosine-protein kinase BTK | | Authors: | Hymowitz, S.G, Maurer, B. | | Deposit date: | 2014-02-13 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent and selective Bruton's tyrosine kinase inhibitors: Discovery of GDC-0834.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
