1LIH
 
 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR | | Authors: | Kim, S.-H, Scott, W, Yeh, J.I, Prive, G.G, Milburn, M. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
4HN4
 
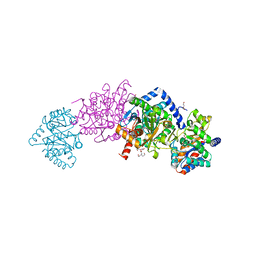 | | Tryptophan synthase in complex with alpha aminoacrylate E(A-A) form and the F9 inhibitor in the alpha site | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BICINE, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2012-10-18 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
6I8L
 
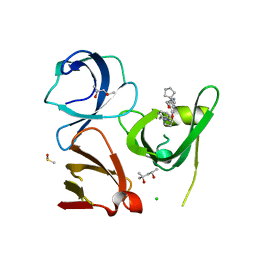 | | Crystal structure of Spindlin1 in complex with the inhibitor TD001851a | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-(cyclopropylmethoxy)-6'-[3-(1,3-dihydroisoindol-2-yl)propoxy]spiro[cyclopentane-1,3'-indole]-2'-amine, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
3WCH
 
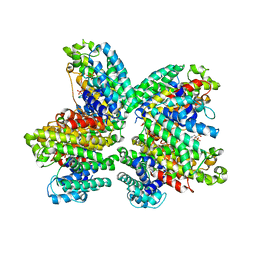 | | The complex structure of HsSQS wtih ligand BPH1237 | | Descriptor: | Squalene synthase, hydrogen [(1R)-2-(3-decyl-1H-imidazol-3-ium-1-yl)-1-hydroxy-1-phosphonoethyl]phosphonate | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
4HNB
 
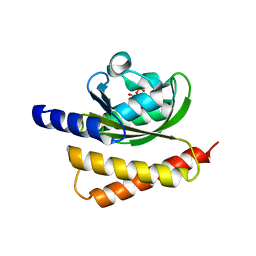 | | Crystal Structure of Rhodobacter Sphaeroides LOV protein | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, FLAVIN MONONUCLEOTIDE, LOV protein | | Authors: | Crane, B.R, Conrad, K.S, Bilwes, A.M. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Light-induced subunit dissociation by a light-oxygen-voltage domain photoreceptor from Rhodobacter sphaeroides.
Biochemistry, 52, 2013
|
|
6CJS
 
 | |
2MUT
 
 | | Solution structure of the F231L mutant ERCC1-XPF dimerization region | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Faridounnia, M, Wienk, H, Kovacic, L, Folkers, G.E, Jaspers, N.G.J, Kaptein, R, Hoeijmakers, J.H.J, Boelens, R. | | Deposit date: | 2014-09-17 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Cerebro-oculo-facio-skeletal Syndrome Point Mutation F231L in the ERCC1 DNA Repair Protein Causes Dissociation of the ERCC1-XPF Complex.
J.Biol.Chem., 290, 2015
|
|
6FT6
 
 | | Structure of the Nop53 pre-60S particle bound to the exosome nuclear cofactors | | Descriptor: | 25S ribosomal RNA, 5S ribosomal RNA, 60S ribosomal protein L11-A, ... | | Authors: | Schuller, J.M, Falk, S, Conti, E. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-28 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the nuclear exosome captured on a maturing preribosome.
Science, 360, 2018
|
|
7GQU
 
 | | Crystal Structure of Werner helicase fragment 517-945 in covalent complex with N-[(E,1S)-1-cyclopropyl-3-methylsulfonylprop-2-enyl]-2-(1,1-difluoroethyl)-4-phenoxypyrimidine-5-carboxamide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, GLYCEROL, ... | | Authors: | Classen, M, Benz, J, Brugger, D, Tagliente, O, Rudolph, M.G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase.
Nature, 629, 2024
|
|
1PC3
 
 | |
2Y7H
 
 | | Atomic model of the DNA-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | 5'-D(*GP*TP*TP*CP*AP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*GP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*CP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*TP*GP*AP*AP*C)-3', S-ADENOSYLMETHIONINE, ... | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
4MAS
 
 | | High Resolution Structure of Wild Type Human Transthyretin in Complex with 3,3',5,5'-tetrachloro-[1,1'-biphenyl]-4,4'diol | | Descriptor: | 3,5,3',5'-TETRACHLORO-BIPHENYL-4,4'-DIOL, Transthyretin | | Authors: | Connelly, S, Bradbury, N.C, Wilson, I.A. | | Deposit date: | 2013-08-16 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | High Resolution Structure of Wild Type Human Transthyretin in Complex with 3,3',5,5'-tetrachloro-[1,1'-biphenyl]-4,4'diol
To be Published
|
|
1Q3X
 
 | | Crystal structure of the catalytic region of human MASP-2 | | Descriptor: | GLYCEROL, Mannan-binding lectin serine protease 2, SODIUM ION | | Authors: | Harmat, V, Gal, P, Kardos, J, Szilagyi, K, Ambrus, G, Naray-Szabo, G, Zavodszky, P. | | Deposit date: | 2003-08-01 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The structure of MBL-associated serine protease-2 reveals that identical substrate specificities of C1s and MASP-2 are realized through different sets of enzyme-substrate interactions
J.Mol.Biol., 342, 2004
|
|
1I0L
 
 | | ANALYSIS OF AN INVARIANT ASPARTIC ACID IN HPRTS-ASPARAGINE MUTANT | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 7-HYDROXY-PYRAZOLO[4,3-D]PYRIMIDINE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, ... | | Authors: | Canyuk, B, Focia, P.J, Eakin, A.E. | | Deposit date: | 2001-01-29 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The role for an invariant aspartic acid in hypoxanthine phosphoribosyltransferases is examined using saturation mutagenesis, functional analysis, and X-ray crystallography.
Biochemistry, 40, 2001
|
|
3L3A
 
 | | Bace-1 with the aminopyridine Compound 32 | | Descriptor: | 4-(4-{1-[(6-aminopyridin-2-yl)methyl]-5-(2-chlorophenyl)-1H-pyrrol-2-yl}phenoxy)butanenitrile, Beta-secretase 1 | | Authors: | Olland, A.M, Chopra, R. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.362 Å) | | Cite: | Novel pyrrolyl 2-aminopyridines as potent and selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OII
 
 | |
9QD7
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 15a | | Descriptor: | (2~{S},9~{S})-2-cyclohexyl-19,22-dimethoxy-11,14,17-trioxa-4-azatricyclo[16.2.2.0^{4,9}]docosa-1(21),18(22),19-triene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Brudy, C, Hausch, F. | | Deposit date: | 2025-03-06 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (2.264 Å) | | Cite: | Linker Modification Enables Control of Key Functional Group Orientation in Macrocycles.
J.Med.Chem., 2025
|
|
4HLB
 
 | |
9QD8
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 29a | | Descriptor: | (2~{S},9~{S},12~{S})-2-cyclohexyl-19,22-dimethoxy-12-methyl-11,14,17-trioxa-4-azatricyclo[16.2.2.0^{4,9}]docosa-1(21),18(22),19-triene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Brudy, C, Hausch, F. | | Deposit date: | 2025-03-06 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Linker Modification Enables Control of Key Functional Group Orientation in Macrocycles.
J.Med.Chem., 2025
|
|
2Y9M
 
 | | Pex4p-Pex22p structure | | Descriptor: | 1,2-ETHANEDIOL, PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into Ubiquitin-Conjugating Enzyme/ Co-Activator Interactions from the Structure of the Pex4P:Pex22P Complex.
Embo J., 31, 2011
|
|
1UDT
 
 | | Crystal structure of Human Phosphodiesterase 5 complexed with Sildenafil(Viagra) | | Descriptor: | 5-{2-ETHOXY-5-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-1-METHYL-3-PROPYL-1H,6H,7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Sung, B.-J, Lee, J.I, Heo, Y.-S, Kim, J.H, Moon, J, Yoon, J.M, Hyun, Y.-L, Kim, E, Eum, S.J, Lee, T.G, Cho, J.M, Park, S.-Y, Lee, J.-O, Jeon, Y.H, Hwang, K.Y, Ro, S. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules
Nature, 425, 2003
|
|
9QD9
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 29c | | Descriptor: | (2~{S},9~{S},12~{S})-2-cyclohexyl-20,23-dimethoxy-12-methyl-11,14,18-trioxa-4-azatricyclo[17.2.2.0^{4,9}]tricosa-1(22),19(23),20-triene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Brudy, C, Hausch, F. | | Deposit date: | 2025-03-06 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Linker Modification Enables Control of Key Functional Group Orientation in Macrocycles.
J.Med.Chem., 2025
|
|
3WQ9
 
 | | Crystal structure of Hsp90-alpha N-terminal domain in complex with 2-(4-Hydroxy-cyclohexylamino)-4-[5-(4-phenyl-imidazol-1-yl)-isoquinolin-1-yl]-benzamide | | Descriptor: | 2-[(trans-4-hydroxycyclohexyl)amino]-4-[5-(4-phenyl-1H-imidazol-1-yl)isoquinolin-1-yl]benzamide, Heat shock protein HSP 90-alpha | | Authors: | Chong, K.T, Yamashita, S, Oshiumi, H, Uno, T, Kitade, M. | | Deposit date: | 2014-01-23 | | Release date: | 2015-02-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of highly selective Hsp90 / inhibitors by structure and thermodynamics guided design
To be Published
|
|
9QDB
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 29d | | Descriptor: | (2~{S},9~{S},12~{R})-2-cyclohexyl-20,23-dimethoxy-12-methyl-11,14,18-trioxa-4-azatricyclo[17.2.2.0^{4,9}]tricosa-1(22),19(23),20-triene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Brudy, C, Hausch, F. | | Deposit date: | 2025-03-06 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Linker Modification Enables Control of Key Functional Group Orientation in Macrocycles.
J.Med.Chem., 2025
|
|
9QDA
 
 | | The FK1 domain of FKBP51 in complex with the macrocyclic SAFit analog 29b | | Descriptor: | (2~{S},9~{S},12~{R})-2-cyclohexyl-19,22-dimethoxy-12-methyl-11,14,17-trioxa-4-azatricyclo[16.2.2.0^{4,9}]docosa-1(21),18(22),19-triene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Brudy, C, Hausch, F. | | Deposit date: | 2025-03-06 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Linker Modification Enables Control of Key Functional Group Orientation in Macrocycles.
J.Med.Chem., 2025
|
|
